| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,161,782 – 14,161,880 |
| Length | 98 |
| Max. P | 0.652059 |

| Location | 14,161,782 – 14,161,880 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.51 |
| Shannon entropy | 0.20522 |
| G+C content | 0.46075 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14161782 98 - 27905053 UACACAAACAAUGGCGGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACAACUCGCACACA- ................(((((((((((....))))..((..((..(((...((((((.....)))))))))..)).))..........)))))))...- ( -25.20, z-score = -2.15, R) >droSim1.chr3R 20200994 98 + 27517382 UACACAAACAAUGGCGGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACAACUCGCACACA- ................(((((((((((....))))..((..((..(((...((((((.....)))))))))..)).))..........)))))))...- ( -25.20, z-score = -2.15, R) >droSec1.super_5 129924 83 + 5866729 UACACAAACAAUGGCGGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACA---------------- ................(((((((((((....))))..........(((...((((((.....)))))))))..)))))))...---------------- ( -23.50, z-score = -2.01, R) >droYak2.chr3R 2772176 98 - 28832112 UACACAAACAAUGGCAGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACAACUCGCACACA- ................(((((((((((....))))..((..((..(((...((((((.....)))))))))..)).))..........)))))))...- ( -25.40, z-score = -2.44, R) >droEre2.scaffold_4770 10270578 96 + 17746568 UACACAAACAAUGGCGGU--GGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACAACUUGCGCACA- ...........(((((((--(((((((....))))..(((((((...)))))))........))))))))))...((((...........))))....- ( -23.10, z-score = -1.04, R) >droAna3.scaffold_13340 13758503 99 + 23697760 UACACAAACAAUGGCGGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACCAACCGCACUACC ............((..((((((.((((....))))..........((((((((((((.....)))))(((......))))).)))))..))))))..)) ( -28.70, z-score = -2.39, R) >dp4.chr2 18746735 97 + 30794189 UACACAAACAAUGGCGGUGC-GGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCACACACAAGCAGCCCCCACAUG- ................(((.-((((((.((.(((...))).))..(((...((((((.....)))))))))...............)))))))))...- ( -24.10, z-score = -0.72, R) >droPer1.super_0 8451421 97 - 11822988 UACACAAACAAUGGCGGUGC-GGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCACACACAAGCAGCCUCCACAUG- ...........(((.(((((-..((((....))))..))......(((...((((((.....)))))))))................))).)))....- ( -21.90, z-score = -0.35, R) >consensus UACACAAACAAUGGCGGUGCGGGGGCUACUAAGCCAAGCUAAGAAGGUUUUGGUAAACAAAUUUUACUGCCAACUCGCACACAAACAACUCGCACACA_ ...........((((((((....((((....))))..(((((((...)))))))..........))))))))........................... (-19.66 = -19.68 + 0.02)


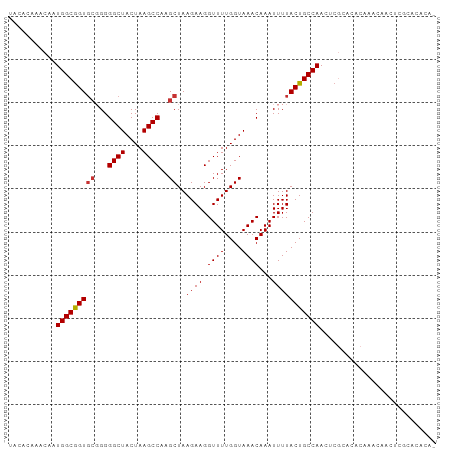
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:25:04 2011