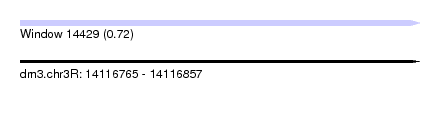
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,116,765 – 14,116,857 |
| Length | 92 |
| Max. P | 0.716873 |

| Location | 14,116,765 – 14,116,857 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 65.94 |
| Shannon entropy | 0.68276 |
| G+C content | 0.62638 |
| Mean single sequence MFE | -38.59 |
| Consensus MFE | -15.65 |
| Energy contribution | -14.13 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14116765 92 + 27905053 -----------ACCGCCGAUGCCAUCGAGGAGUUGGACUUGGUCAUGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGCGGUCGCGGCGGUGGCAUCUUCUU- -----------...((((.((((.((((((.(((((...(((.((.....)).)))....))))).))))))(((((....))))))))).))))........- ( -38.80, z-score = -0.86, R) >droSim1.chr3R 20151920 92 - 27517382 -----------ACCGCCGAUGCCAUCGAGGAGUUGGACUUGGCCAUGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGCGGUCGCGGCGGUGGCAUCUUCUU- -----------...((((.((((.((((((.(((((...(((.((.....)).)))....))))).))))))(((((....))))))))).))))........- ( -38.80, z-score = -0.62, R) >droSec1.super_12 2089366 92 - 2123299 -----------ACCGCAGAUGCCAUCGAGGAGUUGGACUUGGCCAUGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGCGGUCGCGGCGGUGGCAUCUUCUU- -----------...(.((((((((((((((.(((((...(((.((.....)).)))....))))).))))..((.(((.....))).)))))))))))).)..- ( -39.50, z-score = -1.03, R) >droYak2.chr3R 2743680 92 + 28832112 -----------ACCGCCGAUGCCAUCGAGGAGGUGGACUUGGCCACGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGUGGUCGCGGCGGUGGCAUCUUCUU- -----------(((((((..(((((((((((.((((......)))).))))(((((.((.....))...)))))..)))))))..)))))))...........- ( -43.90, z-score = -1.83, R) >droEre2.scaffold_4770 10228622 92 - 17746568 -----------ACCGCCGAUGCCAUCGAGGAGGUGGGCUUGGCCACGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGCGGUCGCGGCGGCGGCAUCUUCUU- -----------...((((.((((.((((((.(.(((...(((.((.....)).)))....))).).))))))(((((....))))))))).))))........- ( -39.30, z-score = 0.32, R) >droPer1.super_3 3984914 94 - 7375914 ---------AUCCACCUGUUGUUGGCC-UUUGCCAGUGUCGUUCUCCUCAUCGUAAUGCUCAUCCGACGACGGCGAUGGCGACGGCAACAGUGGAGGUCCCUUU ---------.(((((.(((((((((((-.(((((..(((((.......(((....)))......)))))..))))).)))..)))))))))))))......... ( -34.62, z-score = -2.02, R) >droWil1.scaffold_181108 3638028 83 + 4707319 ------------CCUCCGG--AUGCC--GCCGCUUC-GUUAG---AACGGUGUCCAUUUACCUGGACUUUUGGUGGCGGCGGCCGCGGCGGUGGCAUCUUCUU- ------------.....((--(((((--((((((((-((...---.)))).(((((......))))).....(((((....))))))))))))))))))....- ( -39.80, z-score = -2.89, R) >droGri2.scaffold_15116 1588558 101 + 1808639 CCACCUCCACCACCCCCGGCAUUGCC--UGUGCUUAAGCUAGCCAGCUUGCGCCCAUUGACUUGCACUUUGGGCGGUGGCGGUCGCGGCGGCGGCAUUUUCUU- ....(.(((((.......(((..((.--((.(((......))))))).)))(((((..(.......)..)))))))))).)(((((....)))))........- ( -36.00, z-score = 0.55, R) >droMoj3.scaffold_6540 12935132 101 + 34148556 CCACCUCCACCACCCCCAAUGCUCCC--UGAGCUCAAGCUAGCCAGUUUACGGCCAUUUAUCUGAACUUUGGGCGGUGGCGGUCGUGGCGGUGGCAUUUUCUU- (((((.((((.(((.(((.(((((..--..(((....))).(((.......)))................))))).))).))).)))).))))).........- ( -36.60, z-score = -1.64, R) >consensus ___________ACCGCCGAUGCCAUCGAGGAGCUGGACUUGGCCACGUUCUGCCCAUUGACCAGCACCUUGGGCGGCGGCGGUCGCGGCGGUGGCAUCUUCUU_ .................(((.(((((........((((........))))((((((.............))))))((.((....)).))))))).)))...... (-15.65 = -14.13 + -1.52)

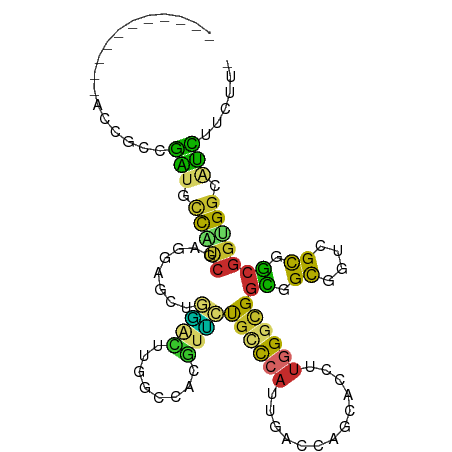
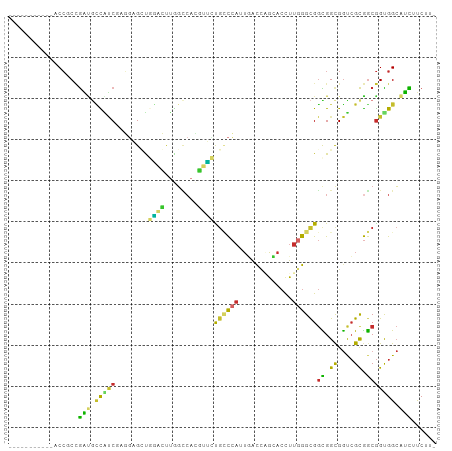
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:58 2011