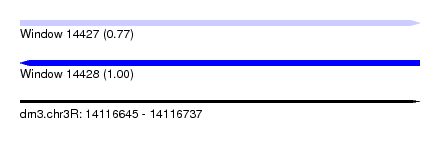
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,116,645 – 14,116,737 |
| Length | 92 |
| Max. P | 0.996032 |

| Location | 14,116,645 – 14,116,737 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.52837 |
| G+C content | 0.58851 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
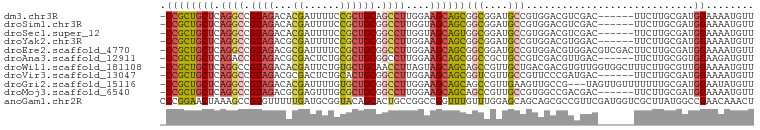
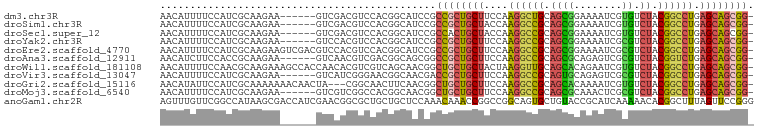
>dm3.chr3R 14116645 92 + 27905053 -CCGCUGCUCAGGCCGUAGACACGAUUUUCCGCUGCAGCCUUGGAAGCAGCGGCGGAUGCCGUGGACGUCGAC------UUCUUGCGAUGGAAAAUGUU -(((((((((((((.((((...((......)))))).))))..).))))))(((....)))..)).(((((.(------.....))))))......... ( -30.80, z-score = 0.10, R) >droSim1.chr3R 20151800 92 - 27517382 -CCGCUGCUCAGGCCGUAGACACGAUUUUCCGCUGCGGCCUUGGUAGCAGCGGCGGAUGCCGUGGACGUCGAC------UUCUUGCGAUGGAAAAUGUU -((((((((.(((((((((...((......))))))))))).)))))).(((((....))))))).(((((.(------.....))))))......... ( -41.30, z-score = -2.72, R) >droSec1.super_12 2089246 92 - 2123299 -CCGCUGCUCAGGCCGUAGACACGAUUUUCCGCUGCGGCCUUGGUAGCAGUGGCGGAUGCCGUGGACGUCGAC------UUCUUGCGAUGGAAAAUGUU -((((((((.(((((((((...((......))))))))))).))))))...(((....)))..)).(((((.(------.....))))))......... ( -37.60, z-score = -1.90, R) >droYak2.chr3R 2743560 92 + 28832112 -CCGCUGCUCAGGCCGUAGACGCGAUUUUCCGCUGCGGCCUUGGAAGCAGCGGCGGAUGCCGUGGACGUGGAC------UUCUUGCGAUGGAAAAUGUU -((((((((((((((((((.((.(.....))))))))))))..).))))))))((....((((.(....((..------..))..).))))....)).. ( -37.80, z-score = -1.33, R) >droEre2.scaffold_4770 10228496 98 - 17746568 -CCGCUGCUCAGGCCGUAGACGCGAUUUUCCGCUGCGGCCUUGGAAGCAGCGGCGGAUGCCGUGGACGUGGACGUCGACUUCUUGCGAUGGAAAAUGUU -.(((((((((((((((((.((.(.....))))))))))))..).)))))))((((...)))).(((((...(((((.(.....))))))....))))) ( -42.20, z-score = -1.76, R) >droAna3.scaffold_12911 3219522 92 - 5364042 -CCGCUGCUCAGACCGUAGACGCGACUCUGCGCUGCGGCCUUGGAAGCAGCGGCCGCUGCCGUCGACGUUGAC------UUCUUGCGGUGGAAGAUGUU -(((((((((((.((((((.((((....)))))))))).))..).))))))))(((((((.((((....))))------.....)))))))........ ( -41.30, z-score = -1.79, R) >droWil1.scaffold_181108 3637908 98 + 4707319 -CCGCUGCUCAGGCCGUAGACACGAUUCUGUGCUGCAACCUUAGUAGCAGCAGCCGUUGCUGACGACGUGUUGGUGGCUUUCUUGCGUUGGAAAAUGUU -((((((((.(((..((((.((((....))))))))..))).)))))).((((..((..(..((.....))..)..))....))))...))........ ( -34.50, z-score = -0.90, R) >droVir3.scaffold_13047 16619292 92 + 19223366 -CCGCUGCUCAGGCCGUAGACGCGACUCUGCACUGCGGCCUUGGAAGCAGCGGUCGUUGCCGUUCCCGAUGAC------UUCUUGCGAUGGAAAAUGUU -((((((((((((((((((..(((....))).)))))))))..).))))))))(((((((.(((......)))------.....)))))))........ ( -37.30, z-score = -2.01, R) >droGri2.scaffold_15116 1588438 95 + 1808639 -CCGCUGCUCAGGCCGUAGACACGAUUUUGUGCUGCGGCCUUGGAAGCAGCAGCCGUUGAAGUUGCCG---UAGUUGUUUUUUUGCGAUGGAAUAUGUU -(((((.((.(((((((((.((((....))))))))))))).)).))).(((((.......)))))((---(((........)))))..))........ ( -36.10, z-score = -2.53, R) >droMoj3.scaffold_6540 12935000 92 + 34148556 -CCGCUGCUCAGGCCGUAGACGCGAGUUUGCGCUGCGGCCUUGGAAGCAGCAGCCGUUGCCGUGGCCGACGAC------UUCUUGCGAUGGAAAAUGUU -..((((((((((((((((.((((....)))))))))))))..).))))))..((((((((((.....)))..------.....)))))))........ ( -40.61, z-score = -1.83, R) >anoGam1.chr2R 603303 99 + 62725911 CCCGGAACUAAAGCCGUGUUUUUGAUGCGGUACAGCACUGCCGGCCGGUUUGUUUGGAGCAGCAGCGCCGUUCGAUGGUCGCUUAUGGCCGAACAAACU .((((.......((((((((...)))))))).........))))..(((((((((((..((..((((((((...)))).))))..)).))))))))))) ( -36.29, z-score = -1.04, R) >consensus _CCGCUGCUCAGGCCGUAGACACGAUUUUGCGCUGCGGCCUUGGAAGCAGCGGCCGAUGCCGUGGACGUCGAC______UUCUUGCGAUGGAAAAUGUU .((((((((.((((.((((...((......)))))).))))....))))))(((....)))............................))........ (-17.23 = -17.53 + 0.29)
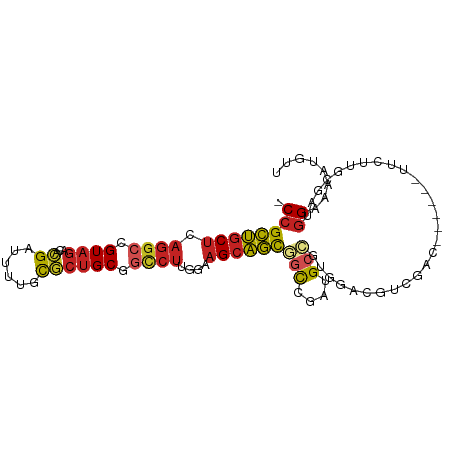
| Location | 14,116,645 – 14,116,737 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.52837 |
| G+C content | 0.58851 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.59 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
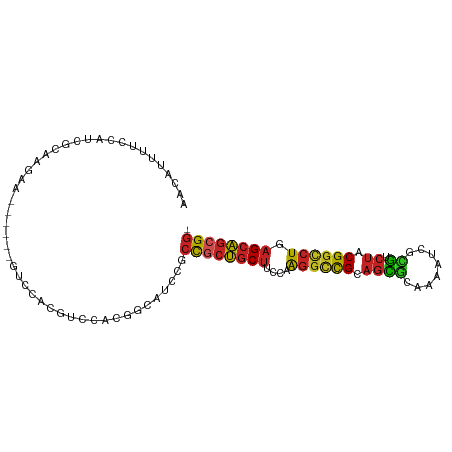
>dm3.chr3R 14116645 92 - 27905053 AACAUUUUCCAUCGCAAGAA------GUCGACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCUGCAGCGGAAAAUCGUGUCUACGGCCUGAGCAGCGG- ...........((....)).------((((.......)))).....(((((((((...((((((..(((......))).....)))))))))))))))- ( -31.90, z-score = -1.29, R) >droSim1.chr3R 20151800 92 + 27517382 AACAUUUUCCAUCGCAAGAA------GUCGACGUCCACGGCAUCCGCCGCUGCUACCAAGGCCGCAGCGGAAAAUCGUGUCUACGGCCUGAGCAGCGG- ...........((....)).------((((.......)))).....((((((((....((((((..(((......))).....)))))).))))))))- ( -34.50, z-score = -2.23, R) >droSec1.super_12 2089246 92 + 2123299 AACAUUUUCCAUCGCAAGAA------GUCGACGUCCACGGCAUCCGCCACUGCUACCAAGGCCGCAGCGGAAAAUCGUGUCUACGGCCUGAGCAGCGG- ...........((....)).------((((.......)))).....((.(((((....((((((..(((......))).....)))))).))))).))- ( -29.10, z-score = -1.08, R) >droYak2.chr3R 2743560 92 - 28832112 AACAUUUUCCAUCGCAAGAA------GUCCACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCCGCAGCGGAAAAUCGCGUCUACGGCCUGAGCAGCGG- ...........((....)).------........((..(((....)))(((((((...((((((.(((((.....).)).)).)))))))))))))))- ( -33.70, z-score = -2.22, R) >droEre2.scaffold_4770 10228496 98 + 17746568 AACAUUUUCCAUCGCAAGAAGUCGACGUCCACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCCGCAGCGGAAAAUCGCGUCUACGGCCUGAGCAGCGG- ........((.((....)).((.((((....)))).))))......(((((((((...((((((.(((((.....).)).)).)))))))))))))))- ( -39.70, z-score = -3.21, R) >droAna3.scaffold_12911 3219522 92 + 5364042 AACAUCUUCCACCGCAAGAA------GUCAACGUCGACGGCAGCGGCCGCUGCUUCCAAGGCCGCAGCGCAGAGUCGCGUCUACGGUCUGAGCAGCGG- ...........((((.....------(((......)))....))))(((((((((...((((((.(((((......))).)).)))))))))))))))- ( -39.30, z-score = -2.31, R) >droWil1.scaffold_181108 3637908 98 - 4707319 AACAUUUUCCAACGCAAGAAAGCCACCAACACGUCGUCAGCAACGGCUGCUGCUACUAAGGUUGCAGCACAGAAUCGUGUCUACGGCCUGAGCAGCGG- ............(....).......((.....(((((.....))))).((((((....((((((.(((((......))).)).)))))).))))))))- ( -30.20, z-score = -1.19, R) >droVir3.scaffold_13047 16619292 92 - 19223366 AACAUUUUCCAUCGCAAGAA------GUCAUCGGGAACGGCAACGACCGCUGCUUCCAAGGCCGCAGUGCAGAGUCGCGUCUACGGCCUGAGCAGCGG- .....(((((.((....)).------(....))))))((....)).(((((((((...((((((.(((((......))).)).)))))))))))))))- ( -39.80, z-score = -3.40, R) >droGri2.scaffold_15116 1588438 95 - 1808639 AACAUAUUCCAUCGCAAAAAAACAACUA---CGGCAACUUCAACGGCUGCUGCUUCCAAGGCCGCAGCACAAAAUCGUGUCUACGGCCUGAGCAGCGG- ............................---.(....)........(((((((((...((((((.(((((......))).)).)))))))))))))))- ( -30.20, z-score = -3.28, R) >droMoj3.scaffold_6540 12935000 92 - 34148556 AACAUUUUCCAUCGCAAGAA------GUCGUCGGCCACGGCAACGGCUGCUGCUUCCAAGGCCGCAGCGCAAACUCGCGUCUACGGCCUGAGCAGCGG- ........((.((....))(------(((((..((....)).))))))(((((((...((((((.(((((......))).)).)))))))))))))))- ( -41.40, z-score = -3.44, R) >anoGam1.chr2R 603303 99 - 62725911 AGUUUGUUCGGCCAUAAGCGACCAUCGAACGGCGCUGCUGCUCCAAACAAACCGGCCGGCAGUGCUGUACCGCAUCAAAAACACGGCUUUAGUUCCGGG ..((((..(((.......((.....)).((((((((((((..((.........)).)))))))))))).)))...))))....(((........))).. ( -28.20, z-score = 0.47, R) >consensus AACAUUUUCCAUCGCAAGAA______GUCCACGUCCACGGCAUCCGCCGCUGCUUCCAAGGCCGCAGCGCAAAAUCGCGUCUACGGCCUGAGCAGCGG_ ..............................................((((((((....((((((.((((........)).)).)))))).)))))))). (-22.86 = -22.59 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:57 2011