
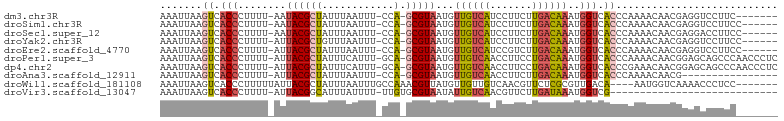
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,085,699 – 14,085,792 |
| Length | 93 |
| Max. P | 0.871254 |

| Location | 14,085,699 – 14,085,792 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Shannon entropy | 0.44512 |
| G+C content | 0.38413 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.49 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14085699 93 + 27905053 AAAUUAAGUCACCCUUUU-AAUACGCUAUUUAAUUU-CCA-GCGUAAUGUUGUCAUCCUUCUUGACAAAUGGUCACCCAAAACAACGAGGUCCUUC------- ............((((..-..((((((.........-..)-))))).((((((((.......))))...(((....))).))))..))))......------- ( -15.30, z-score = -1.72, R) >droSim1.chr3R 20121729 94 - 27517382 AAAUUAAGUCACCCUUUU-AAUACGCUAUUUAAUUU-CCA-GCGUAAUGUUGUCAUCCUUCUUGACAAAUGGUCACCCAAAACAACGAGGUCCUUCC------ ............((((..-..((((((.........-..)-))))).((((((((.......))))...(((....))).))))..)))).......------ ( -15.30, z-score = -1.70, R) >droSec1.super_12 2058500 94 - 2123299 AAAUUAAGUCACCCUUUU-AAUACGCUAUUUAAUUU-CCA-GCGUAAUGUUGUCAUCCUUCUUGACAAAUGGUCACCCAAAACAACGAGGACCUUCC------ ............((((..-..((((((.........-..)-))))).((((((((.......))))...(((....))).))))..)))).......------ ( -15.80, z-score = -1.68, R) >droYak2.chr3R 2694705 94 + 28832112 AAAUUAAGUCACCCUUUU-AUUACGCUGUUUAAUUU-CCA-GCGUAAUGUUGUCAUCCUUCUUGACAAAUGGUCACCCAAAACAACGAGGUCCUUCC------ ............((((.(-(((((((((........-.))-))))))))((((((.......)))))).(((....))).......)))).......------ ( -20.70, z-score = -3.35, R) >droEre2.scaffold_4770 10196878 94 - 17746568 AAAUUAAGUCACCCUUUU-AUUACGCUAUUUAAUUU-CCA-GCGUAAUGUUGUCAUCCGUCUUGACAAAUGGUCACCCAAAACAACGAGGUCCUUCC------ ............((((.(-((((((((.........-..)-))))))))((((((.......)))))).(((....))).......)))).......------ ( -17.70, z-score = -2.32, R) >droPer1.super_3 5257511 100 - 7375914 AAAUUAAGUCACCCUUUU-AUUACGCUAUUUCAUUU-GCA-GCGUAAUGUUGUCAACCUUCCUGACAAAUGGUCACCCAAAACAACGGAGCAGCCCAACCCUC .......((.(((....(-((((((((.........-..)-))))))))((((((.......))))))..))).))..........((......))....... ( -17.80, z-score = -1.55, R) >dp4.chr2 11423219 100 - 30794189 AAAUUAAGUCACCCUUUU-AUUACGCUAUUUCAUUU-GCA-GCGUAAUGUUGUCAACCUUCCUGACAAAUGGUCACCCGAAACAACGGAGCAGCCCAACCCUC .................(-((((((((.........-..)-))))))))((((((.......))))))..(((..((((......))).)..)))........ ( -19.30, z-score = -1.79, R) >droAna3.scaffold_12911 3187420 84 - 5364042 AAAUUAAGUCACCCUUUU-AUUACGCUAUUUAAUUU-CCA-GCGUAAUGUUGUCAACCUUCUUGACAAAUGGUCACCCAAAACAACG---------------- .......((.(((....(-((((((((.........-..)-))))))))(((((((.....)))))))..))).))...........---------------- ( -17.30, z-score = -4.19, R) >droWil1.scaffold_181108 1033518 92 - 4707319 AAAUUAAGUCACCCUUUUUAUUACGCUAUUUAAUUUGCCAAACGUUAUGUUGUUGUCAACGUUCUCGCGUUGACA----AAUGGUCAAAACCCUCC------- .................................((((((((((.....))).((((((((((....)))))))))----).))).)))).......------- ( -17.30, z-score = -2.35, R) >droVir3.scaffold_13047 19177651 73 + 19223366 AAAUUAAGUCACCCUUUU-AUUACGGCAUUUAUUUU-UUGUGCGUAAUAUUGUCAACGUUCUUGAUAAAUGGUCG---------------------------- ..........(((....(-((((((.(((.......-..))))))))))(((((((.....)))))))..)))..---------------------------- ( -12.50, z-score = -1.43, R) >consensus AAAUUAAGUCACCCUUUU_AUUACGCUAUUUAAUUU_CCA_GCGUAAUGUUGUCAUCCUUCUUGACAAAUGGUCACCCAAAACAACGAGGUCCUUC_______ .......((.(((........(((((...............)))))...((((((.......))))))..))).))........................... ( -7.85 = -8.49 + 0.64)


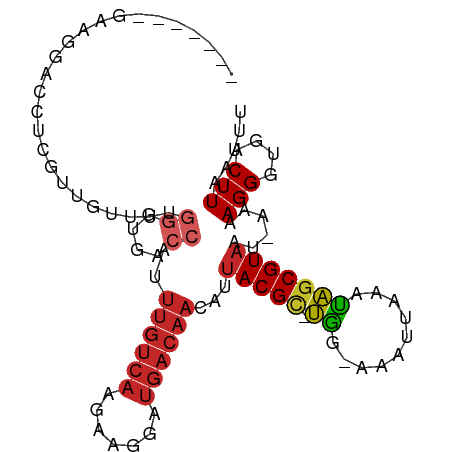
| Location | 14,085,699 – 14,085,792 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Shannon entropy | 0.44512 |
| G+C content | 0.38413 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -10.26 |
| Energy contribution | -12.31 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14085699 93 - 27905053 -------GAAGGACCUCGUUGUUUUGGGUGACCAUUUGUCAAGAAGGAUGACAACAUUACGC-UGG-AAAUUAAAUAGCGUAUU-AAAAGGGUGACUUAAUUU -------.(((.((((.((((((.(((....)))...(((......)))))))))..(((((-((.-........)))))))..-....))))..)))..... ( -23.70, z-score = -2.23, R) >droSim1.chr3R 20121729 94 + 27517382 ------GGAAGGACCUCGUUGUUUUGGGUGACCAUUUGUCAAGAAGGAUGACAACAUUACGC-UGG-AAAUUAAAUAGCGUAUU-AAAAGGGUGACUUAAUUU ------..(((.((((.((((((.(((....)))...(((......)))))))))..(((((-((.-........)))))))..-....))))..)))..... ( -23.70, z-score = -2.08, R) >droSec1.super_12 2058500 94 + 2123299 ------GGAAGGUCCUCGUUGUUUUGGGUGACCAUUUGUCAAGAAGGAUGACAACAUUACGC-UGG-AAAUUAAAUAGCGUAUU-AAAAGGGUGACUUAAUUU ------...((((((((((((((.(((....)))...(((......)))))))))..(((((-((.-........)))))))..-....))).)))))..... ( -27.00, z-score = -2.99, R) >droYak2.chr3R 2694705 94 - 28832112 ------GGAAGGACCUCGUUGUUUUGGGUGACCAUUUGUCAAGAAGGAUGACAACAUUACGC-UGG-AAAUUAAACAGCGUAAU-AAAAGGGUGACUUAAUUU ------..(((.((((.((((((.(((....)))...(((......)))))))))(((((((-((.-........)))))))))-....))))..)))..... ( -28.50, z-score = -3.61, R) >droEre2.scaffold_4770 10196878 94 + 17746568 ------GGAAGGACCUCGUUGUUUUGGGUGACCAUUUGUCAAGACGGAUGACAACAUUACGC-UGG-AAAUUAAAUAGCGUAAU-AAAAGGGUGACUUAAUUU ------..(((.((((.((((((..((....))((((((....))))))))))))(((((((-((.-........)))))))))-....))))..)))..... ( -27.60, z-score = -3.32, R) >droPer1.super_3 5257511 100 + 7375914 GAGGGUUGGGCUGCUCCGUUGUUUUGGGUGACCAUUUGUCAGGAAGGUUGACAACAUUACGC-UGC-AAAUGAAAUAGCGUAAU-AAAAGGGUGACUUAAUUU ..((((((((..((.((.......(((....))).(((((((.....))))))).(((((((-((.-........)))))))))-....))))..)))))))) ( -27.90, z-score = -1.66, R) >dp4.chr2 11423219 100 + 30794189 GAGGGUUGGGCUGCUCCGUUGUUUCGGGUGACCAUUUGUCAGGAAGGUUGACAACAUUACGC-UGC-AAAUGAAAUAGCGUAAU-AAAAGGGUGACUUAAUUU ..((((((((..((.((........((....))..(((((((.....))))))).(((((((-((.-........)))))))))-....))))..)))))))) ( -27.50, z-score = -1.40, R) >droAna3.scaffold_12911 3187420 84 + 5364042 ----------------CGUUGUUUUGGGUGACCAUUUGUCAAGAAGGUUGACAACAUUACGC-UGG-AAAUUAAAUAGCGUAAU-AAAAGGGUGACUUAAUUU ----------------.......((((((.(((..(((((((.....))))))).(((((((-((.-........)))))))))-.....))).))))))... ( -24.80, z-score = -3.98, R) >droWil1.scaffold_181108 1033518 92 + 4707319 -------GGAGGGUUUUGACCAUU----UGUCAACGCGAGAACGUUGACAACAACAUAACGUUUGGCAAAUUAAAUAGCGUAAUAAAAAGGGUGACUUAAUUU -------...(((((....((..(----((((((((......))))))))).......(((((.............)))))........))..)))))..... ( -18.62, z-score = -1.12, R) >droVir3.scaffold_13047 19177651 73 - 19223366 ----------------------------CGACCAUUUAUCAAGAACGUUGACAAUAUUACGCACAA-AAAAUAAAUGCCGUAAU-AAAAGGGUGACUUAAUUU ----------------------------..(((.....((((.....))))...(((((((((...-........)).))))))-)....))).......... ( -8.50, z-score = -0.22, R) >consensus _______GAAGGACCUCGUUGUUUUGGGUGACCAUUUGUCAAGAAGGAUGACAACAUUACGC_UGG_AAAUUAAAUAGCGUAAU_AAAAGGGUGACUUAAUUU ................((((((((.((....))..((((((.......))))))..................))))))))........(((....)))..... (-10.26 = -12.31 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:55 2011