| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,065,009 – 14,065,095 |
| Length | 86 |
| Max. P | 0.980968 |

| Location | 14,065,009 – 14,065,095 |
|---|---|
| Length | 86 |
| Sequences | 9 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 54.32 |
| Shannon entropy | 0.93926 |
| G+C content | 0.48216 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -1.12 |
| Energy contribution | -0.84 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 2.20 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.04 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14065009 86 + 27905053 --AACGGGUGACGAGUGACCUCCCUCCAAAUUGGAAAGCUUCGUUGCCCAACAAACUUAGUUUUAGUUAU--UCUACUCGUAGACUAAGU --...(((..((((((....(((.........)))..)).))))..))).....(((((((((.(((...--...)))...))))))))) ( -25.90, z-score = -3.15, R) >droVir3.scaffold_13047 24641 85 - 19223366 -----CAGCGGCCAGCAGCAUGAGGCAAAAUCCAAUUUGGUUUUGUCUCAACUCUGGCCCAAUAAUUAAUCGCCUAGUUGUAUGUAUAAA -----..(.((((((.((..((((((((((((......)))))))))))).))))))))).(((((((......)))))))......... ( -29.30, z-score = -4.18, R) >droSim1.chr3R 20101437 85 - 27517382 --AACGGGUGACGAGUGACCUCCCUCCCA-CCGGGACACUUCGUUGCCCAACAAACUUAGUUUUAGUUAU--CCUACUCGUAGGCUAAGU --...(((..(((((((...((((.....-..))))))).))))..))).....(((((((((.(((...--...)))...))))))))) ( -31.20, z-score = -3.66, R) >droSec1.super_12 2037823 87 - 2123299 AACACGGGUGACGAGUGACCUCCCUCCCA-CUGGGACACUUCGUUGCCCAACAAACUUAGUUUUAGUUAU--CCUACUCGUAGGCUAAGU .....(((..(((((((...((((.....-..))))))).))))..))).....(((((((((.(((...--...)))...))))))))) ( -31.80, z-score = -3.80, R) >droYak2.chr3R 2673858 80 + 28832112 ---ACGGGAGACAAGUGACCUCCCAC-----UGGGAAGCUUAGUCGCCCAACAAAUUUAGUUCUAGUUAU--CCUACUCGUAGGCUAAGC ---..(((.(((((((....((((..-----.)))).)))).))).))).................(((.--((((....)))).))).. ( -21.00, z-score = -0.77, R) >droEre2.scaffold_4770 10176368 80 - 17746568 ---ACGGGAGACGAGUGACUUCCCCC-----UGGGAAGCUUCGUCGCCCAACAAACUUAGCUCUAGUUAU--CCUACUCGUAGGCUAAGC ---..(((.((((((.(.((((((..-----.))))))))))))).)))......((((((((.(((...--...))).)..))))))). ( -31.00, z-score = -3.21, R) >droAna3.scaffold_12911 3167476 76 - 5364042 ---AUGGAUACAGGGCGGCUCUUCCG-----CAAAGAGCCCCGGAUCCGUACUACUUUAAGUAUAUCCCUACACCAUUGCAAAA------ ---((((......((.(((((((...-----..)))))))))((((..(((((......))))))))).....)))).......------ ( -22.60, z-score = -2.43, R) >droPer1.super_6 1670802 76 - 6141320 ---ACGGGCGACGGAAGAGCUAC----------AGAUAGCUCGUCCGUCCCCAAAAACAAAUUUAGCCAUAGCCUAGCCCUAAGUUAAA- ---..(((.((((((.((((((.----------...)))))).))))))))).......(((((((.............)))))))...- ( -27.12, z-score = -4.86, R) >dp4.chr2 1647693 76 - 30794189 ---ACGGGCGACGGAAGAGCUAC----------AGAUAGCUCGUCCGUCCCCAAAAACAAAUUUAGCCAUAGCCUAGCCCUAAGUUAAA- ---..(((.((((((.((((((.----------...)))))).))))))))).......(((((((.............)))))))...- ( -27.12, z-score = -4.86, R) >consensus ___ACGGGUGACGAGUGACCUCCCUC_____UGGGAAACUUCGUCGCCCAACAAACUUAGUUUUAGUUAU__CCUACUCGUAGGCUAAG_ .....((..(((((.....(((..........))).....)))))..))......................................... ( -1.12 = -0.84 + -0.28)

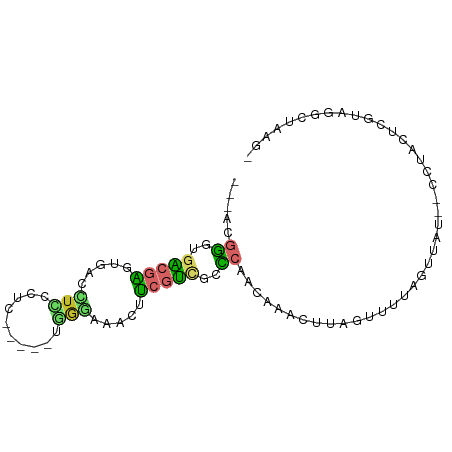
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:51 2011