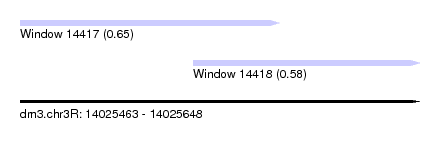
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,025,463 – 14,025,648 |
| Length | 185 |
| Max. P | 0.650680 |

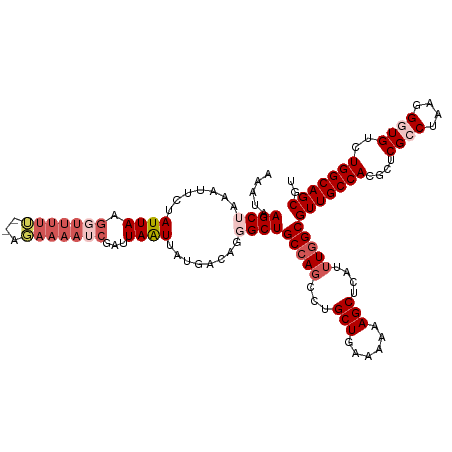
| Location | 14,025,463 – 14,025,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Shannon entropy | 0.26416 |
| G+C content | 0.41887 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -23.56 |
| Energy contribution | -24.44 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14025463 120 + 27905053 AAAUGAGCUAAAUUCUAUUAAGGUUUAAUUAUAUUAUCGAUUAAUUAUGACAGGGCUGCCAGCGUGCUGGAAAAAGCUCAUUUGGCGUUGCCACGGUCGCCUAAGGGUGUCUGGCAGCGU (((((((((............(((((..((((((((.....))).)))))..))))).((((....))))....))))))))).(((((((((.((.((((....))))))))))))))) ( -40.70, z-score = -2.77, R) >droSim1.chr3R 20061709 118 - 27517382 AAAUAAGCAAAAUUCUAUUAAGGUUUUU--AGAAAAUCGAUUAAUUAUGACAGAGCUGCCAGCCUGCUGAAAAAAGCUCAUUUGGCGUUGCCACGGUCGCCUAAAGGUGUCUGGCAGCGU .......((((.(((((..........)--))))..................(((((..(((....))).....))))).))))(((((((((.((.((((....))))))))))))))) ( -33.50, z-score = -1.52, R) >droSec1.super_12 1996672 118 - 2123299 AAAUAAGCUAAAUACUAUUAAGGUUUUA--AGAAAAUCGAUUAAUUAUGACAGGGCUGCCAGCCUGCUGAAAAAAGCUCAUUUGGCGUUGCCACGAUCGCCUAAGGGUGUCUGGCAGCGU ......(((((((........(((((..--...)))))..............(((((..(((....))).....))))))))))))(((((((.((.((((....))))))))))))).. ( -35.70, z-score = -1.92, R) >droYak2.chr3R 2627274 118 + 28832112 AUAAAAGCUGAUUUCUAUUAAAUAUUUU--AGAAAAUCGAUUAGUUAAUGCAAGGCUGCCAUCCUGCUUAAAAAAGCUCGCUUUGCGUUGCCAGGCUCGCCCAAGGUUGUCUGGCAGCGU .....((((((((((((..........)--))).....))))))))...(((((((.........((((....))))..)))))))((((((((((..(((...))).)))))))))).. ( -36.10, z-score = -2.23, R) >droEre2.scaffold_4770 10136429 118 - 17746568 AUAAAAGCAAAAUUCUAUUAUGUUUUUU--AGAAAAUCGAUUAAUUACGACAAGGCUGCCAGCUUGCUCAAAAAAGCUCAUUUGGCGUUGCCAGGCUCGUCCAAGGUUGUCUGGCAGCGU .....(((....(((((..(....)..)--))))..(((........)))....)))(((((...(((......)))....)))))((((((((((....(....)..)))))))))).. ( -32.20, z-score = -1.47, R) >consensus AAAUAAGCUAAAUUCUAUUAAGGUUUUU__AGAAAAUCGAUUAAUUAUGACAGGGCUGCCAGCCUGCUGAAAAAAGCUCAUUUGGCGUUGCCACGCUCGCCUAAGGGUGUCUGGCAGCGU .....((((.......(((((....(((.....)))....)))))........))))(((((...(((......)))....)))))(((((((....((((....))))..))))))).. (-23.56 = -24.44 + 0.88)

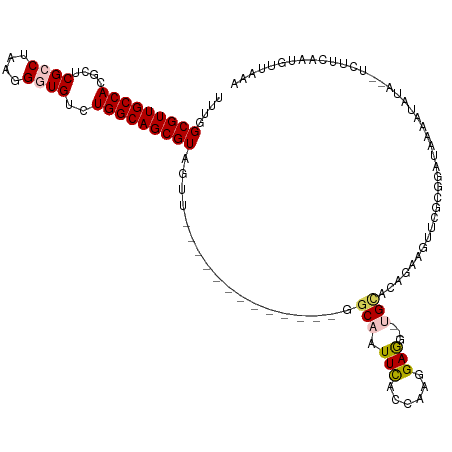
| Location | 14,025,543 – 14,025,648 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Shannon entropy | 0.39618 |
| G+C content | 0.46219 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 14025543 105 + 27905053 UUUGGCGUUGCCACGGUCGCCUAAGGGUGUCUGGCAGCGUAGUU-------------GGCAAUUCACCAAGGAGGACCGUACAGAAGUUCGCGGAUAAAAUAUA--UCUUAAAUGCUAAA .((((((((...((((((.(((...((((..((.((((...)))-------------).))...)))).)))..))))))............(((((.....))--)))..)))))))). ( -33.30, z-score = -1.73, R) >droSim1.chr3R 20061787 102 - 27517382 UUUGGCGUUGCCACGGUCGCCUAAAGGUGUCUGGCAGCGUAGUU-------------GGCAAUUUACCAAGGAG---UGCACAGGAGUUCGCGGAUAAAUUAUA--UAUUUAAUGUUAAA ....(((((((((.((.((((....)))))))))))))))..((-------------((((............(---((.((....)).)))...(((((....--.))))).)))))). ( -27.60, z-score = -0.68, R) >droSec1.super_12 1996750 102 - 2123299 UUUGGCGUUGCCACGAUCGCCUAAGGGUGUCUGGCAGCGUAGUU-------------GGCAAUUCACCAAGGAG---CGCACAGAAGUUCGCGAAUAAAAUAUA--UCUUUCGUGUUAAA ....(((((((((.((.((((....)))))))))))))))..((-------------(((...........(((---(........))))(((((.........--...)))))))))). ( -31.40, z-score = -1.57, R) >droYak2.chr3R 2627352 116 + 28832112 CUUUGCGUUGCCAGGCUCGCCCAAGGUUGUCUGGCAGCGUUGCCCAUGUU-UCGUA-GUCAAUUCGCCAAGGAAG--UGCAUAGAAGUUCGCUGAUAAAGUAAAUUUCUUCAAUAUUACU ....((((((((((((..(((...))).))))))))))))..........-..(((-((......(((......)--.))..(((((((..((.....))..))))))).....))))). ( -33.10, z-score = -1.86, R) >droEre2.scaffold_4770 10136507 118 - 17746568 UUUGGCGUUGCCAGGCUCGUCCAAGGUUGUCUGGCAGCGUAGCCAUUGGUAUCGUAUGGCAAUUCGCCAAGGAAG--UGCGUAGAAGUUCGCGGAUAAAAUAUACUUCUUCAAUAAUUCA ((((((((((((((((....(....)..))))))))))...(((((((....)).))))).....))))))(((.--.....((((((...............)))))).......))). ( -37.78, z-score = -2.31, R) >consensus UUUGGCGUUGCCACGCUCGCCUAAGGGUGUCUGGCAGCGUAGUU_____________GGCAAUUCACCAAGGAGG__UGCACAGAAGUUCGCGGAUAAAAUAUA__UCUUCAAUGUUAAA ....(((((((((....((((....))))..)))))))))..................(((.(((......)))...)))........................................ (-18.20 = -18.68 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:49 2011