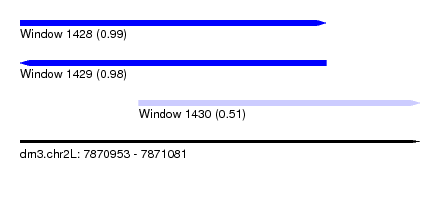
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,870,953 – 7,871,081 |
| Length | 128 |
| Max. P | 0.991932 |

| Location | 7,870,953 – 7,871,051 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Shannon entropy | 0.12474 |
| G+C content | 0.40292 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.94 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
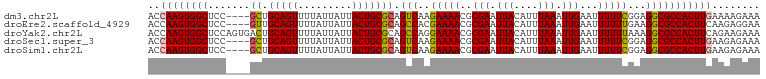
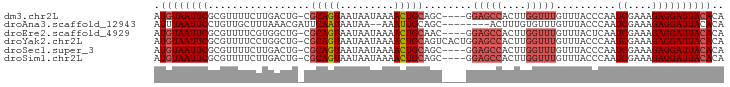
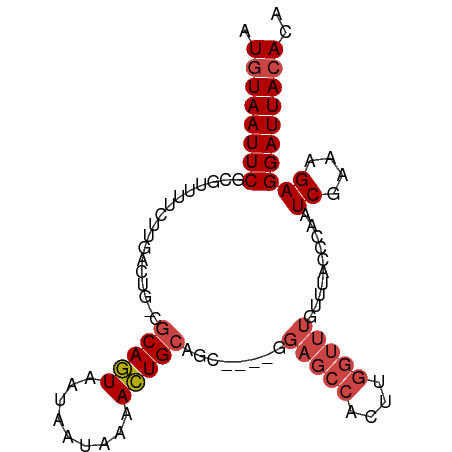
>dm3.chr2L 7870953 98 + 23011544 UUUCUUUUCAAGUGGCGCCUCCGAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCUUGACUGCGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGU .......(((((((((.............((((...((((((.....))))))..))))(((((((((.........))))).))----)).))))))))). ( -30.60, z-score = -3.67, R) >droEre2.scaffold_4929 16792771 98 + 26641161 UUCCUCUUGAAGUGGCGCCUUCAAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCGUGGCUGCGCAGUAAUAAUAAAACUGCAAC----GGAGCCACUUGGU .......(.(((((((.((.....................(((..(((((....))))).)))(((((.........)))))...----)).))))))).). ( -26.10, z-score = -2.03, R) >droYak2.chr2L 17299555 102 - 22324452 UUUCUUCUGAAGUGGCGCCUUUAAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCCUGGCUGCGCAGUAAUAAUAAAACUGCAGUCACUGGAGCCACUUGGU .........(((((((....................((((((.....))))))((((((((((..(((.........))))))))))..))))))))))... ( -25.20, z-score = -1.59, R) >droSec1.super_3 3384946 98 + 7220098 UUUCUCUUCAAGUGGCGCCUCCGAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCUUGACUGCGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGU .......(((((((((.............((((...((((((.....))))))..))))(((((((((.........))))).))----)).))))))))). ( -30.60, z-score = -3.76, R) >droSim1.chr2L 7668497 98 + 22036055 UUUCUCUUCAAGUGGCGCCUCCGAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCUUGACUGCGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGU .......(((((((((.............((((...((((((.....))))))..))))(((((((((.........))))).))----)).))))))))). ( -30.60, z-score = -3.76, R) >consensus UUUCUCUUCAAGUGGCGCCUCCGAAAAAUUCAAUUUAAAUGUAAUUCGCGUUUUCUUGACUGCGCAGUAAUAAUAAAACUGCAGC____GGAGCCACUUGGU .......(((((((((.((..........((((...((((((.....))))))..))))....(((((.........))))).......)).))))))))). (-25.38 = -25.94 + 0.56)

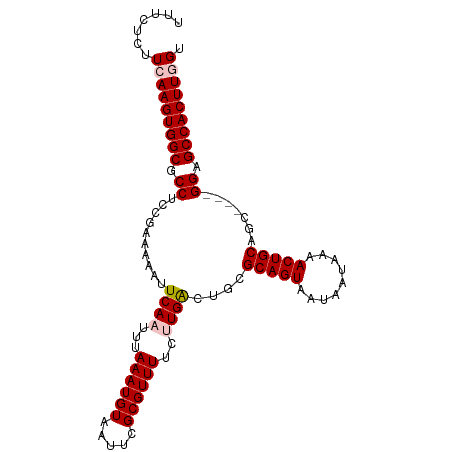
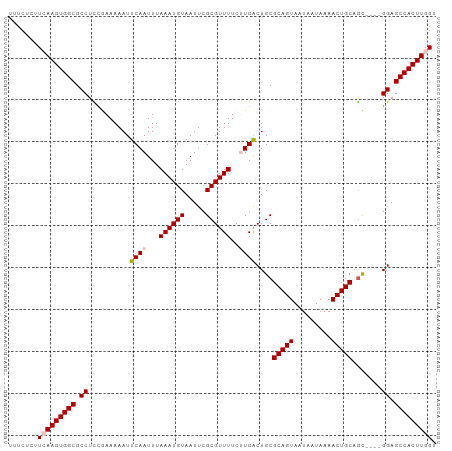
| Location | 7,870,953 – 7,871,051 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Shannon entropy | 0.12474 |
| G+C content | 0.40292 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7870953 98 - 23011544 ACCAAGUGGCUCC----GCUGCAGUUUUAUUAUUACUGCGCAGUCAAGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUCGGAGGCGCCACUUGAAAAGAAA ..((((((((.((----((.(((((.........)))))))..((..(((((..(((.(((....))).)))..)))))..)))).))))))))........ ( -29.90, z-score = -3.26, R) >droEre2.scaffold_4929 16792771 98 - 26641161 ACCAAGUGGCUCC----GUUGCAGUUUUAUUAUUACUGCGCAGCCACGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUUGAAGGCGCCACUUCAAGAGGAA .((..((((((..----((.(((((.........))))))))))))).............................((((((((......)))))))))).. ( -26.00, z-score = -2.12, R) >droYak2.chr2L 17299555 102 + 22324452 ACCAAGUGGCUCCAGUGACUGCAGUUUUAUUAUUACUGCGCAGCCAGGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUUAAAGGCGCCACUUCAGAAGAAA ...(((((((.((.((((..(((((.........)))))((............))...)))).((((((........)))))))).)))))))......... ( -23.70, z-score = -1.30, R) >droSec1.super_3 3384946 98 - 7220098 ACCAAGUGGCUCC----GCUGCAGUUUUAUUAUUACUGCGCAGUCAAGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUCGGAGGCGCCACUUGAAGAGAAA ..((((((((.((----((.(((((.........)))))))..((..(((((..(((.(((....))).)))..)))))..)))).))))))))........ ( -29.90, z-score = -3.10, R) >droSim1.chr2L 7668497 98 - 22036055 ACCAAGUGGCUCC----GCUGCAGUUUUAUUAUUACUGCGCAGUCAAGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUCGGAGGCGCCACUUGAAGAGAAA ..((((((((.((----((.(((((.........)))))))..((..(((((..(((.(((....))).)))..)))))..)))).))))))))........ ( -29.90, z-score = -3.10, R) >consensus ACCAAGUGGCUCC____GCUGCAGUUUUAUUAUUACUGCGCAGUCAAGAAAACGCGAAUUACAUUUAAAUUGAAUUUUUCGGAGGCGCCACUUGAAGAGAAA ..((((((((.......((.(((((.........))))))).(((..(((((..(((.(((....))).)))...)))))...)))))))))))........ (-25.08 = -25.04 + -0.04)
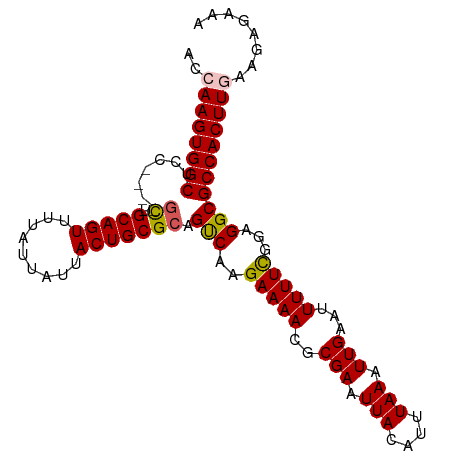
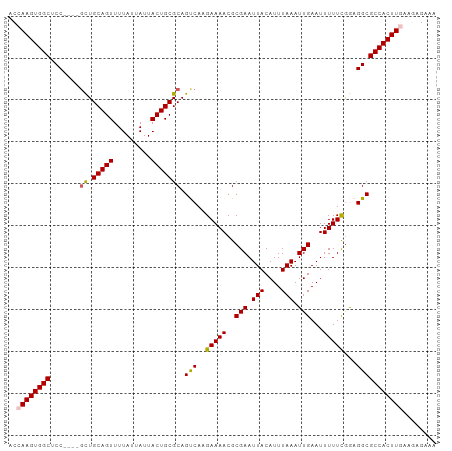
| Location | 7,870,991 – 7,871,081 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Shannon entropy | 0.24180 |
| G+C content | 0.39598 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7870991 90 + 23011544 AUGUAAUUCGCGUUUUCUUGACUG-CGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .((((((((...(((((....(((-((((((.........))))).))----))((((....)))).............))))).)))))))).. ( -24.80, z-score = -2.20, R) >droAna3.scaffold_12943 3604719 85 - 5039921 AUUUAAUUCCUGUUGCUUUAAACGAUUCAAUAAUAA--AAAUUGCAGC--------ACUUUGUGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .......(((((((((....................--.....)))))--------.((((.((.(((......))).))))))))))....... ( -10.91, z-score = 0.14, R) >droEre2.scaffold_4929 16792809 90 + 26641161 AUGUAAUUCGCGUUUUCGUGGCUG-CGCAGUAAUAAUAAAACUGCAAC----GGAGCCACUUGGUUUGUUUACUCAAUCGAAAGAGGAUUACACA .((((((((...(((((((((((.-((((((.........)))))...----).))))))(((((......)).)))..))))).)))))))).. ( -28.30, z-score = -3.12, R) >droYak2.chr2L 17299593 94 - 22324452 AUGUAAUUCGCGUUUUCCUGGCUG-CGCAGUAAUAAUAAAACUGCAGUCACUGGAGCCACUUGGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .(((((((((.(((((..((((((-(..(((.........))))))))))..))))))....(((......)))...((....)))))))))).. ( -24.90, z-score = -1.51, R) >droSec1.super_3 3384984 90 + 7220098 AUGUAAUUCGCGUUUUCUUGACUG-CGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .((((((((...(((((....(((-((((((.........))))).))----))((((....)))).............))))).)))))))).. ( -24.80, z-score = -2.20, R) >droSim1.chr2L 7668535 90 + 22036055 AUGUAAUUCGCGUUUUCUUGACUG-CGCAGUAAUAAUAAAACUGCAGC----GGAGCCACUUGGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .((((((((...(((((....(((-((((((.........))))).))----))((((....)))).............))))).)))))))).. ( -24.80, z-score = -2.20, R) >consensus AUGUAAUUCGCGUUUUCUUGACUG_CGCAGUAAUAAUAAAACUGCAGC____GGAGCCACUUGGUUUGUUUACCCAAUCGAAAGAGGAUUACACA .((((((((.................(((((.........)))))........(((((....)))))..........((....)))))))))).. (-15.71 = -16.60 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:35 2011