
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 14,014,950 – 14,015,051 |
| Length | 101 |
| Max. P | 0.607388 |

| Location | 14,014,950 – 14,015,051 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 52.21 |
| Shannon entropy | 1.07239 |
| G+C content | 0.49233 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -8.89 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.91 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
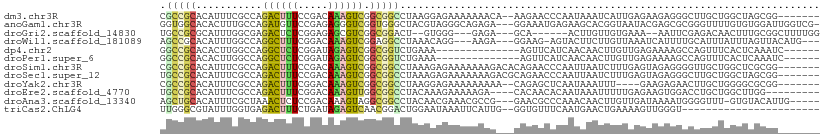
>dm3.chr3R 14014950 101 - 27905053 CGCCGCACAUUUCGCCAGACUUUCCGACAAAGUCGGCGGCCUAAGGAGAAAAAAACA--AAGAACCCAAUAAAUCAUUGAGAAGAGGGCUUGCUGGCUAGCGG------- .((((((.....((((.((((((.....))))))))))((((..((...........--.....))((((.....))))......)))).))).)))......------- ( -24.49, z-score = 0.04, R) >anoGam1.chr3R 25541643 106 + 53272125 GGUGGCACACUUUGCCAGAUGUUCCGAGAGGGUCGGUGGGCUACGUAGGGCAGAGA---GGAAAUGAGAAGCACGGUAAUACGAGCGCGGGUUUUGUGUGGAUUGGUCG- ..(.((((.(((((((..((((.((....))(((....))).))))..))))))).---.(((((..(..((.((......)).)).)..))))))))).)........- ( -29.30, z-score = 0.04, R) >droGri2.scaffold_14830 4872956 94 + 6267026 UGCCGCGCAUUUGGCGAGACUCUCGGAGAGCGUCGGCGGACU--GUGGG---GAGA---GCA------ACUUGUUGUGAAA--AAUUCGAGACAACUUUGCGGCUUUUGG ..((.((((((((.(((..(((.....)))..))).)))).)--))).)---)(((---((.------.(..(((((....--........)))))...)..)))))... ( -27.70, z-score = 0.57, R) >droWil1.scaffold_181089 7663705 100 + 12369635 AGCCGCACAUUUGGCCAGGCUUUCGGACAAAGUCGGAGGCCUAAACAGG---AAGA---GGAAG-AGUACUUCUUGUUAAAUCAUUUUGCAUUUAUUUAGUUACAUG--- .((((......)))).((((((((((......)))))))))).((((((---(((.---.....-....))))))))).............................--- ( -25.90, z-score = -1.71, R) >dp4.chr2 29105773 90 - 30794189 GGCCGCACACUUGGCCAGGCUCUCGGAUAGAGUCGGCGGUCUGAAA--------------AGUUCAUCAACAACUUGUUGAGAAAAGCCAGUUUCACUCAAAUC------ (((((......))))).((((((.....))))))((((..((....--------------))..).(((((.....))))).....)))...............------ ( -27.10, z-score = -1.75, R) >droPer1.super_6 4430082 90 - 6141320 GGCCGCACACUUGGCCAGGCUCUCGGAUAGAGUCGGCGGUCUGAAA--------------AGUUCAUCAACAACUUGUUGAGAAAAGCCAGUUUCACUCAAAUC------ (((((......))))).((((((.....))))))((((..((....--------------))..).(((((.....))))).....)))...............------ ( -27.10, z-score = -1.75, R) >droSim1.chr3R 20051331 103 + 27517382 CGCCGCACAUUUCGCCAGACUUUCCGACAAAGUCGGCGGCCUAAAGAGAAAAAAAGACACAGAACCCAAUUAAUCUUUGAGUAGAGGGGUUGCUGGCUCGCGG------- ..((((.....(((((.((((((.....)))))))))))................((..((((((((......(((......))).))))).)))..))))))------- ( -31.80, z-score = -1.62, R) >droSec1.super_12 1986198 103 + 2123299 UGCCGCACAUUUCGCCAGACUUUCCGACAAAGUCGGCGGCCUAAAGAGAAAAAAAGACGCAGAACCCAAUUAAUCUUUGAGUAGAGGGCUUGCUGGCUAGCGG------- ..((((......((((.((((((.....))))))))))(((((((((..........................))))))(((((.....))))))))..))))------- ( -26.17, z-score = 0.29, R) >droYak2.chr3R 2616917 97 - 28832112 CGCCGCACAUUUCGCCAGACUUUCGGACAAAGUCGGCGGCCUAAGGAGAAAAAAAAA--CAGAGCUCAAUAAAUUU----GAAGAGAACUUGCUGGGGCGCGG------- ..((((......((((.((((((.....))))))))))((((.(((((.........--.....))).........----.(((....))).)).))))))))------- ( -26.34, z-score = -0.61, R) >droEre2.scaffold_4770 10126006 97 + 17746568 UGCCGCACAUUUCGCCAGACUUUCGGACAAAGUUGGCGGCCUACAAAGAAAAAGA----CACAACACAAUAAAUUUUUGAGAAGUGGACCUGCUGGCUUGG--------- .(((((((((((((((((.((((.....)))))))))......((((((......----..............)))))).))))))....))).)))....--------- ( -21.55, z-score = -0.15, R) >droAna3.scaffold_13340 15782973 101 - 23697760 AGCUGCACAUUUCGCUAAACUCUCCGACAAAGUAGGCGGCCUACAACGAAACGCCG---GAACGCCCAAACAACUUGUUGAUAAAAUGGGGUUU-GUGUACAUUG----- .(.((((((..((.(((.......((((((.((.((((.............))))(---(.....)).....))))))))......)))))..)-))))))....----- ( -21.84, z-score = 0.40, R) >triCas2.ChLG4 11503110 85 + 13894384 UUGGGCGUAUUUGGUGAGACUUUCUGAUAGAGUCAACGGACUGGAAUAAAUUCAUUG--GGUGUUUCAAUGAACUGAAAAGUUGGGU----------------------- ...((..((((..(((((...((((.....((((....))))))))....)))))..--))))..))..(.((((....)))).)..----------------------- ( -17.50, z-score = -0.45, R) >consensus UGCCGCACAUUUCGCCAGACUUUCGGACAAAGUCGGCGGCCUAAAAAGAAAAAAAA___GAGAACACAAUUAAUUUUUAAAUAAAGGCCUGGCUGGCUCGCGG_______ .(((((...........((((((.....)))))).)))))...................................................................... ( -8.89 = -8.68 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:46 2011