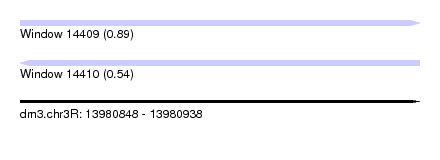
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,980,848 – 13,980,938 |
| Length | 90 |
| Max. P | 0.890078 |

| Location | 13,980,848 – 13,980,938 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 74.60 |
| Shannon entropy | 0.53833 |
| G+C content | 0.48685 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.71 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.890078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13980848 90 + 27905053 CCCUGCAUUUCUCACUCCGC--CGAUUGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC--GCUGGAGAAAGAGAAUAGUGCUGACGAUCG ....(((((((((.((((((--.((((((...(.(((((.....))))).)....)))))).--)).))))...)))))..))))......... ( -27.00, z-score = -1.95, R) >droEre2.scaffold_4770 10090859 85 - 17746568 CCCUGCAUUUCUCACUCCGC--CGAUGGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC--GCUGGAGAAAGAGAACAA-GCGAAUG---- ..((((((((((.....(((--.(.....).)))(((((.....)))))))))))))))(((--(((..............)-)))))..---- ( -23.24, z-score = -1.11, R) >droYak2.chr3R 2577393 85 + 28832112 CCCUGCAUUUCUCACUCCGC--CGAUUGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC--GCUGGAGAAAGAGAAUAA-GAGAAUG---- ..((....(((((.((((((--.((((((...(.(((((.....))))).)....)))))).--)).))))...)))))..)-)......---- ( -23.60, z-score = -2.08, R) >droSec1.super_12 1951153 90 - 2123299 CCCUGCAUUUCUCACUCCGC--CGAUUGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC--GCUGGAGAAAGAGAAUAGUGCUGCAGAUCG ..(((((.(((((.((((((--.((((((...(.(((((.....))))).)....)))))).--)).))))...)))))......))))).... ( -28.40, z-score = -2.07, R) >droSim1.chr3R 20015464 90 - 27517382 CCCUGCAUUUCUCACUCCGC--CGAUUGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC--GCUGGAGAAAGAGAAUAGUGCUGCCGAUCG ....(((((((((.((((((--.((((((...(.(((((.....))))).)....)))))).--)).))))...)))))..))))......... ( -27.00, z-score = -1.71, R) >dp4.chr2 29071928 91 + 30794189 CCCUGCAUUUCUCAUUCGUU--CGAUUGCAAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUCGCAGUGGUUCGU-UGAGGAGCCAAGAAGGAGG (((((((((((.((((.(((--((((((....))).))))...)).)))))))))))))(((....((((((..-....)))))).)))))... ( -28.50, z-score = -1.81, R) >droPer1.super_6 4396134 91 + 6141320 CCCUGCAUUUCUCAUUCGUU--CGAUUGCAAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUCGCAGUGGUUCGU-UGAGGAGCCAAGAAGGAGG (((((((((((.((((.(((--((((((....))).))))...)).)))))))))))))(((....((((((..-....)))))).)))))... ( -28.50, z-score = -1.81, R) >droAna3.scaffold_13340 11953743 82 - 23697760 CCCUGCAUUUCUCACUCUCCUGCGAUUGCUGGCGAUUUGAUAAGCAAAUGGAAAUGCAGA----GCCGGAGAAAGAG--CAGCGCCGA------ ..(.((.((.(((..(((((.((..((((...(.(((((.....))))).)....)))).----)).)))))..)))--.)).)).).------ ( -23.70, z-score = -0.22, R) >droMoj3.scaffold_6540 31628137 88 - 34148556 CCCUGCAUUUCU--UUUGCA--CGAU-GCAAGCGAUUUGAUAAGCAAAUGGAAAUGCAGCUCCAACUGGGCCAA-UGCAGGCCGCAUCCAGAUG ..((((((((((--(((((.--....-)))))..(((((.....))))))))))))))).......((((((..-....)))).))........ ( -25.60, z-score = -0.53, R) >droVir3.scaffold_13047 12545885 79 - 19223366 CCCUGCAUUUCU--UUUGCA--CGAU-GCAAGCGAUUUGAUAAGCAAAUGGAAAUGCAGCUCCAACUGGGUGAACUGCCAGACA---------- ..((((((((((--(((((.--....-)))))..(((((.....)))))))))))))))......((((........))))...---------- ( -22.60, z-score = -1.49, R) >consensus CCCUGCAUUUCUCACUCCGC__CGAUUGCCAGCGAUUUGAUAAGCAAAUGGAAAUGCAGUUC__GCUGGAGAAAGAGAAUAGCGCAGAAG___G ..((((((((((..........((........))(((((.....)))))))))))))))................................... (-13.71 = -13.71 + -0.00)



| Location | 13,980,848 – 13,980,938 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 74.60 |
| Shannon entropy | 0.53833 |
| G+C content | 0.48685 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13980848 90 - 27905053 CGAUCGUCAGCACUAUUCUCUUUCUCCAGC--GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCAAUCG--GCGGAGUGAGAAAUGCAGGG .........(((...(((((...(((((((--(((.((......)))))))).........((((.....))--)))))).))))).))).... ( -23.20, z-score = -0.48, R) >droEre2.scaffold_4770 10090859 85 + 17746568 ----CAUUCGC-UUGUUCUCUUUCUCCAGC--GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCCAUCG--GCGGAGUGAGAAAUGCAGGG ----..(((((-(.(..........).)))--)))((((((((((((((...........(((((.....))--)))))))).))))))))).. ( -26.50, z-score = -1.81, R) >droYak2.chr3R 2577393 85 - 28832112 ----CAUUCUC-UUAUUCUCUUUCUCCAGC--GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCAAUCG--GCGGAGUGAGAAAUGCAGGG ----((((.((-(((((((((....(((((--((...(((........)))........))))))).....)--).)))))))))))))..... ( -22.70, z-score = -1.48, R) >droSec1.super_12 1951153 90 + 2123299 CGAUCUGCAGCACUAUUCUCUUUCUCCAGC--GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCAAUCG--GCGGAGUGAGAAAUGCAGGG ...((((((......(((((...(((((((--(((.((......)))))))).........((((.....))--)))))).))))).)))))). ( -26.70, z-score = -1.39, R) >droSim1.chr3R 20015464 90 + 27517382 CGAUCGGCAGCACUAUUCUCUUUCUCCAGC--GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCAAUCG--GCGGAGUGAGAAAUGCAGGG .........(((...(((((...(((((((--(((.((......)))))))).........((((.....))--)))))).))))).))).... ( -23.20, z-score = -0.13, R) >dp4.chr2 29071928 91 - 30794189 CCUCCUUCUUGGCUCCUCA-ACGAACCACUGCGAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUUGCAAUCG--AACGAAUGAGAAAUGCAGGG .........(((.((....-..)).))).......(((((((((((((((.((..(((.....))).....)--).)))))).))))))))).. ( -19.20, z-score = -1.14, R) >droPer1.super_6 4396134 91 - 6141320 CCUCCUUCUUGGCUCCUCA-ACGAACCACUGCGAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUUGCAAUCG--AACGAAUGAGAAAUGCAGGG .........(((.((....-..)).))).......(((((((((((((((.((..(((.....))).....)--).)))))).))))))))).. ( -19.20, z-score = -1.14, R) >droAna3.scaffold_13340 11953743 82 + 23697760 ------UCGGCGCUG--CUCUUUCUCCGGC----UCUGCAUUUCCAUUUGCUUAUCAAAUCGCCAGCAAUCGCAGGAGAGUGAGAAAUGCAGGG ------....(.(((--(..((((((..((----((((((........)))...........((.((....)).))))))))))))).)))).) ( -22.50, z-score = 0.32, R) >droMoj3.scaffold_6540 31628137 88 + 34148556 CAUCUGGAUGCGGCCUGCA-UUGGCCCAGUUGGAGCUGCAUUUCCAUUUGCUUAUCAAAUCGCUUGC-AUCG--UGCAAA--AGAAAUGCAGGG ..((..(.((.((((....-..)))))).)..)).(((((((((..(((((..(((((.....))).-))..--.)))))--.))))))))).. ( -29.20, z-score = -0.81, R) >droVir3.scaffold_13047 12545885 79 + 19223366 ----------UGUCUGGCAGUUCACCCAGUUGGAGCUGCAUUUCCAUUUGCUUAUCAAAUCGCUUGC-AUCG--UGCAAA--AGAAAUGCAGGG ----------..((..((..........))..)).(((((((((..(((((..(((((.....))).-))..--.)))))--.))))))))).. ( -20.10, z-score = -0.14, R) >consensus C___CGUCAGCGCUAUUCUCUUUCUCCAGC__GAACUGCAUUUCCAUUUGCUUAUCAAAUCGCUGGCAAUCG__GCGGAGUGAGAAAUGCAGGG ...................................((((((((((((((............................))))).))))))))).. (-13.32 = -13.32 + 0.00)

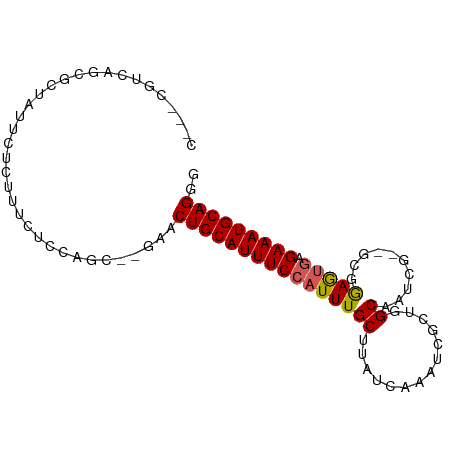
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:42 2011