| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,967,946 – 13,968,044 |
| Length | 98 |
| Max. P | 0.655508 |

| Location | 13,967,946 – 13,968,044 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.63730 |
| G+C content | 0.42631 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
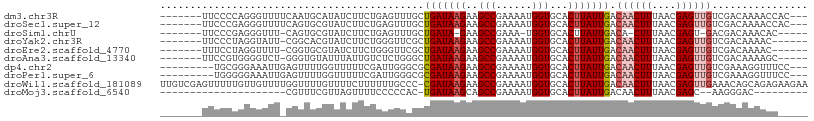
>dm3.chr3R 13967946 98 + 27905053 -------UUCCCCAGGGUUUUCAAUGCAUAUCUUCUGAGUUUGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAACCAC--- -------........((((((....(((..((....))...))).((((((..(((......)))...))))))((((((((....))))))))...))))))..--- ( -24.50, z-score = -1.92, R) >droSec1.super_12 1933612 98 - 2123299 -------UUCCCGAGGGUUUUCAGUGCGUAUCUUCUGAGUUUGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAACCAC--- -------........((((((.(((((...((((.(.((....)).).)))).(((......))))))))..((((((((((....)))))))))).))))))..--- ( -28.10, z-score = -2.27, R) >droSim1.chrU 4289297 91 - 15797150 -------UUCCCGAGGGUUU-CAGUGCGUAUCUUCUGAGUUUGCUGAUA-GAAGCCGAAA-UGGUGCACUUAUUGACA-CUUUAACGAGU-GACGACAAACAC----- -------........(((((-(...(((..((....))...))).....-))))))....-((.((((((..((((..-..))))..)))-).)).)).....----- ( -17.60, z-score = 0.88, R) >droYak2.chr3R 2565043 94 + 28832112 -------UUCCCUAGGUAUU-CGGCACGUAUCUUCUGGGUUCGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAAC------ -------.....(((((...-..((((.(((.(((.((.(((........))).))))).))))))))))))((((((((((....))))))))))......------ ( -25.10, z-score = -1.66, R) >droEre2.scaffold_4770 10078444 94 - 17746568 -------UUUCCUAGGUUUU-CGGUGCGUAUCUUCUGGGUUCGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAAC------ -------........(((((-(((((((.(((.....))).))).........)))))))))..........((((((((((....))))))))))......------ ( -24.50, z-score = -1.30, R) >droAna3.scaffold_13340 11942594 95 - 23697760 -------UUCCGUGGGGUCU-GGGUGUAUUUAUUGUCUCUGGGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAAGC----- -------.........((((-(((((((((.(((.((....((((.......)))))).)))))))))))))..((((((((....)))))))))))......----- ( -26.70, z-score = -1.65, R) >dp4.chr2 29056017 96 + 30794189 ---------UGCGGGAAAUUGAGUUUUGGUUUUUCGAUUGGGCGCGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGAAAGGUUUCC--- ---------....((((((....(((((((((((..((((....)))).)))))))))))........(((.((((((((((....)))))))))))))))))))--- ( -27.30, z-score = -1.80, R) >droPer1.super_6 4380708 96 + 6141320 ---------UGGGGGAAAUUGAGUUUUGGUUUUUCGAUUGGGCGCGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGAAAGGUUUCC--- ---------....((((((....(((((((((((..((((....)))).)))))))))))........(((.((((((((((....)))))))))))))))))))--- ( -27.30, z-score = -2.04, R) >droWil1.scaffold_181089 4746991 107 + 12369635 UUGUCGAGUUUUUGUUGUUUUGGUUUUGUUUUCUUUUUUGCCC-CGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGAAACAGCAGAGAAGAA ...((...((((((((((((.(((...............))).-(((((((..(((......)))...))))))).((((((....)))))))))))))))))).)). ( -26.06, z-score = -1.57, R) >droMoj3.scaffold_6540 31610027 75 - 34148556 ---------------------CGUUUCGUUAGUUUUCCCCCAC-UGAUAAGCAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGC--AAGGGAC--------- ---------------------.(((.((((((((.((......-.((((((..(((......)))...)))))))).))))..)))))))--.......--------- ( -12.51, z-score = 0.58, R) >consensus _______UUCCCGAGGGUUU_CGGUGCGUAUCUUCUGUGUUCGCUGAUAAGAAGCCGAAAAUGGUGCACUUAUUGACAACUUUAACGAGUUGUCGACAAAACC_____ ............................................(((((((..(((......)))...))))))).((((((....))))))................ ( -9.56 = -9.95 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:38 2011