| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,966,411 – 13,966,524 |
| Length | 113 |
| Max. P | 0.942634 |

| Location | 13,966,411 – 13,966,515 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.03 |
| Shannon entropy | 0.54654 |
| G+C content | 0.38842 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -11.24 |
| Energy contribution | -10.21 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
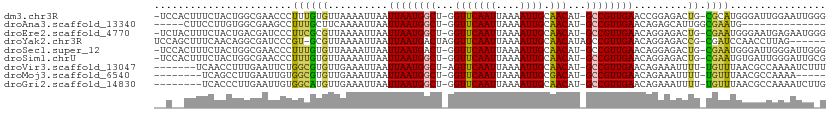
>dm3.chr3R 13966411 104 + 27905053 -GAAACUCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACCGGAGACUG-CGCAUGGGAUU -....(.(((........))).)..(((..(((((.........((((((((.-.(((((((....)))).)))..-))))))))...(....).)-)))).)))... ( -25.30, z-score = -1.57, R) >droAna3.scaffold_13340 11941496 91 - 23697760 ----------CUUCCUUGUGGCGAAGCCUUUGCUUCAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAGCAUUGGCGAAUG----- ----------.(((((....(((((((....)))))........((((((((.-.(((((((....)))).)))..-)))))))).....))...)).)))..----- ( -22.70, z-score = -0.99, R) >droEre2.scaffold_4770 10077124 104 - 17746568 -GAAAGUCUACUUUCUACUGACGAUCCCUUCGCGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGGGAAU -(((((....))))).........(((((((((...........((((((((.-.(((((((....)))).)))..-))))))))..((....)))-)))).)))).. ( -32.80, z-score = -4.43, R) >droYak2.chr3R 2563692 106 + 28832112 GAAAAUCCAGCUUUCAACAGGCGAUCCCGU-GCGUUAAAAUUAAUUAAUGACUAGGUUCAAUUAAAAUUGCAACAUAGCCGUUGAACAGGAGACCG-CGAUCCAACCU ............((((((.(((......((-.((((((......))))))))...(((((((....)))).)))...)))))))))..(((.....-...)))..... ( -19.70, z-score = -0.51, R) >droSec1.super_12 1932338 104 - 2123299 -GAAACUCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGACU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGGGAUU -.....(((...(((....((((((((....(((((((......))))).)).-)))))(((....))).......-)))......(((....)))-.)))..))).. ( -19.50, z-score = -0.48, R) >droSim1.chrU 7267799 104 - 15797150 -GAAACUCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGUGAUU -.......(((.(((.....((((.....))))...........((((((((.-.(((((((....)))).)))..-)))))))).(((....)))-.))).)))... ( -22.30, z-score = -1.26, R) >droVir3.scaffold_13047 12530223 98 - 19223366 --GGACUU---CAACCUUUGA--AUUCUGGCGUGUUGAAAUUAAUUAAUGGCU-AGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCC --(((.((---((.....)))--).)))(((((((((...(((((((((....-.))).)))))).....))))))-)))((((((((((....))-))))))))... ( -28.00, z-score = -3.80, R) >droMoj3.scaffold_6540 31609133 101 - 34148556 --GGACUUGGCUCAGCCUUGA--AUUGUGGCGUGUUGAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCGACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCC --......(((..........--.....(((((((((.((((..(((((....-......))))))))).))))))-)))((((((((((....))-))))))))))) ( -28.10, z-score = -2.14, R) >droGri2.scaffold_14830 3696004 96 - 6267026 ---GACUU----CACCCUUGA--AUUGUGGCAUGUUGAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCC ---...((----((....)))--)....(((((((((.((((..(((((....-......))))))))).))))))-)))((((((((((....))-))))))))... ( -25.40, z-score = -2.79, R) >consensus __AAACUC_ACUUUCUACUGGCGAUCCCUUCGUGUUAAAAUUAAUUAAUGGCU_GGUUCAAUUAAAAUUGCAACAU_GCCGUUGAACAGGAGACUG_CGAAUGAGACU ............................((((............((((((((...(((((((....)))).)))...))))))))............))))....... (-11.24 = -10.21 + -1.03)


| Location | 13,966,416 – 13,966,524 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.58403 |
| G+C content | 0.38693 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -11.24 |
| Energy contribution | -10.21 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13966416 108 + 27905053 -UCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACCGGAGACUG-CGCAUGGGAUUGGAAUUGGG -.(((..((((((....)..(((..(((((.........((((((((.-.(((((((....)))).)))..-))))))))...(....).)-)))).)))..))))).))). ( -27.10, z-score = -0.88, R) >droAna3.scaffold_13340 11941496 91 - 23697760 -----CUUCCUUGUGGCGAAGCCUUUGCUUCAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAGCAUUGGCGAAUG-------------- -----.(((((....(((((((....)))))........((((((((.-.(((((((....)))).)))..-)))))))).....))...)).)))..-------------- ( -22.70, z-score = -0.99, R) >droEre2.scaffold_4770 10077129 108 - 17746568 -UCUACUUUCUACUGACGAUCCCUUCGCGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGGGAAUGAGAAUGGG -.(((.((((..(....).(((((((((...........((((((((.-.(((((((....)))).)))..-))))))))..((....)))-)))).))))..)))).))). ( -30.10, z-score = -2.91, R) >droYak2.chr3R 2563697 104 + 28832112 UCCAGCUUUCAACAGGCGAUCCCGU-GCGUUAAAAUUAAUUAAUGACUAGGUUCAAUUAAAAUUGCAACAUAGCCGUUGAACAGGAGACCG-CGAUCCAACCUUAG------ .......((((((.(((......((-.((((((......))))))))...(((((((....)))).)))...)))))))))..(((.....-...)))........------ ( -19.70, z-score = -0.66, R) >droSec1.super_12 1932343 108 - 2123299 -UCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGACU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGGGAUUGGGAUUGGG -.(((..(((((.....(((((....(((((((......))))).)).-)))))...............((-.(((((...(((....)))-..))))).))))))).))). ( -22.50, z-score = 0.02, R) >droSim1.chrU 7267804 108 - 15797150 -UCCACUUUCUACUGGCGAACCCUUUGUGUUAAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGGAGACUG-CGAAUGUGAUUGGGAUUGCG -..............((((.(((.(..((((........((((((((.-.(((((((....)))).)))..-)))))))).(((....)))-..))))..)..))).)))). ( -29.20, z-score = -2.18, R) >droVir3.scaffold_13047 12530228 102 - 19223366 -------UCAACCUUUGAAUUCUGGCGUGUUGAAAUUAAUUAAUGGCU-AGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCCAAAAUCUUU -------......((((......(((((((((...(((((((((....-.))).)))))).....))))))-)))((((((((((....))-))))))))..))))...... ( -26.00, z-score = -3.32, R) >droMoj3.scaffold_6540 31609142 96 - 34148556 --------UCAGCCUUGAAUUGUGGCGUGUUGAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCGACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCCAAAA----- --------...((..........(((((((((.((((..(((((....-......))))))))).))))))-)))((((((((((....))-)))))))))).....----- ( -25.40, z-score = -2.15, R) >droGri2.scaffold_14830 3696008 101 - 6267026 --------UCACCCUUGAAUUGUGGCAUGUUGAAAUUAAUUAAUGGCU-GGUUCAAUUAAAAUUGCAACAU-GCCGUUGAACAGAAAUUUU-UGUUUAACGCCAAAAUCUUG --------......(((......(((((((((.((((..(((((....-......))))))))).))))))-)))((((((((((....))-))))))))..)))....... ( -25.40, z-score = -2.56, R) >consensus __C__CUUUCUACUGGCGAUCCCUUCGUGUUAAAAUUAAUUAAUGGCU_GGUUCAAUUAAAAUUGCAACAU_GCCGUUGAACAGGAGACUG_CGAAUGAGACUGAAAUUG_G .......................((((............((((((((...(((((((....)))).)))...))))))))............))))................ (-11.24 = -10.21 + -1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:37 2011