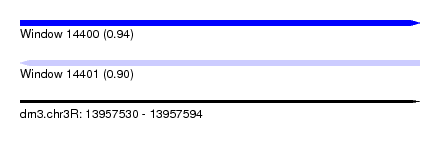
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,957,530 – 13,957,594 |
| Length | 64 |
| Max. P | 0.941375 |

| Location | 13,957,530 – 13,957,594 |
|---|---|
| Length | 64 |
| Sequences | 4 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.45513 |
| G+C content | 0.67458 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
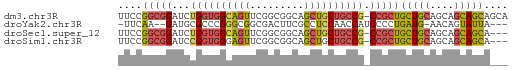
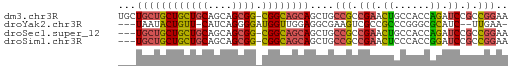
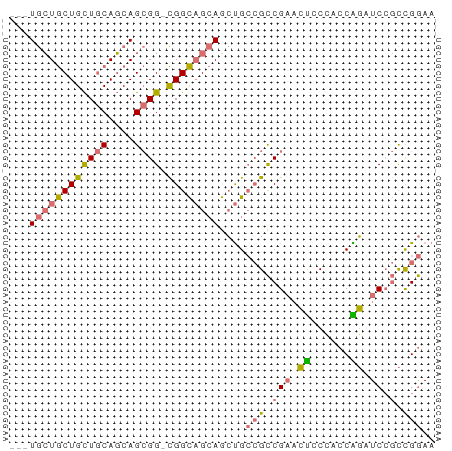
>dm3.chr3R 13957530 64 + 27905053 UUCCGGCGGAUCUGGUGGCAGUUCGGCGGCAGCUGCUGCCG-CCGCUGCUGCAGCAGCAGCAGCA ....((((((.(((....))))))((((((....)))))))-))((((((((....)))))))). ( -37.60, z-score = -1.89, R) >droYak2.chr3R 27532911 58 + 28832112 -UUCAA--GAUGCGCCCGGGCGGCGACUUCGCCUCCAACCAUCCCCUGAUG-AACAGUAUUA--- -.....--(((((....(((.((((....)))))))...((((....))))-....))))).--- ( -16.60, z-score = -1.65, R) >droSec1.super_12 1923501 61 - 2123299 UUCCGGCGGAUCUGGUGGCAGUUCGGCGGCAGCUGCUGCCG-CCGCUGCUGCAGCAGCAGCA--- ....((((((.(((....))))))((((((....)))))))-))(((((((...))))))).--- ( -32.60, z-score = -0.90, R) >droSim1.chr3R 19991594 61 - 27517382 UUCCGGCGGAUCCGGUGGGAGUUCGGCGGCAGCUGCUGCCG-CCGCUGCUGCAGCAGCAGCA--- (((((.((....)).)))))....((((((((...))))))-))(((((((...))))))).--- ( -36.30, z-score = -2.26, R) >consensus UUCCGGCGGAUCCGGUGGCAGUUCGGCGGCAGCUGCUGCCG_CCGCUGCUGCAGCAGCAGCA___ ....(((((...((((((((((.........)))))))))).)))))(((((....))))).... (-20.15 = -20.77 + 0.63)
| Location | 13,957,530 – 13,957,594 |
|---|---|
| Length | 64 |
| Sequences | 4 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.45513 |
| G+C content | 0.67458 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
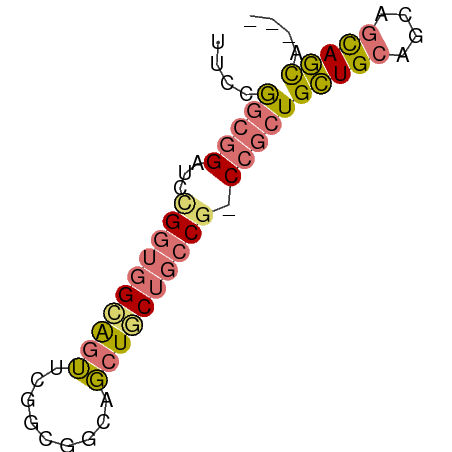
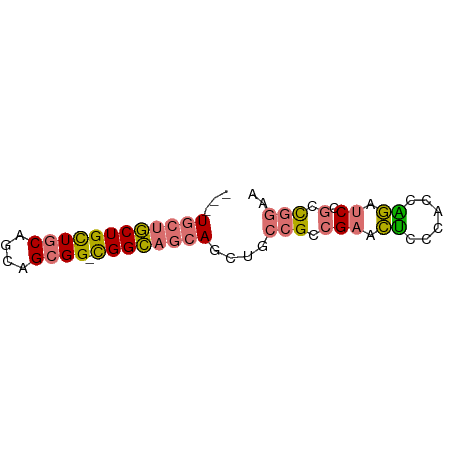
>dm3.chr3R 13957530 64 - 27905053 UGCUGCUGCUGCUGCAGCAGCGG-CGGCAGCAGCUGCCGCCGAACUGCCACCAGAUCCGCCGGAA .((((((((((((((....))))-))))))))))..((((.((.(((....))).)).).))).. ( -36.30, z-score = -2.75, R) >droYak2.chr3R 27532911 58 - 28832112 ---UAAUACUGUU-CAUCAGGGGAUGGUUGGAGGCGAAGUCGCCGCCCGGGCGCAUC--UUGAA- ---..........-..((((((..((.((((.((((....))))..)))).))..))--)))).- ( -20.60, z-score = -1.26, R) >droSec1.super_12 1923501 61 + 2123299 ---UGCUGCUGCUGCAGCAGCGG-CGGCAGCAGCUGCCGCCGAACUGCCACCAGAUCCGCCGGAA ---((((((((((((....))))-))))))))....((((.((.(((....))).)).).))).. ( -29.90, z-score = -1.32, R) >droSim1.chr3R 19991594 61 + 27517382 ---UGCUGCUGCUGCAGCAGCGG-CGGCAGCAGCUGCCGCCGAACUCCCACCGGAUCCGCCGGAA ---((((((....)))))).(((-((((((...)))))))))........((((.....)))).. ( -34.10, z-score = -2.89, R) >consensus ___UGCUGCUGCUGCAGCAGCGG_CGGCAGCAGCUGCCGCCGAACUCCCACCAGAUCCGCCGGAA ...((((((....))))))((((.((((.((....)).))))..((......))..))))..... (-15.11 = -14.93 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:35 2011