| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,947,766 – 13,947,856 |
| Length | 90 |
| Max. P | 0.952419 |

| Location | 13,947,766 – 13,947,856 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.71186 |
| G+C content | 0.48236 |
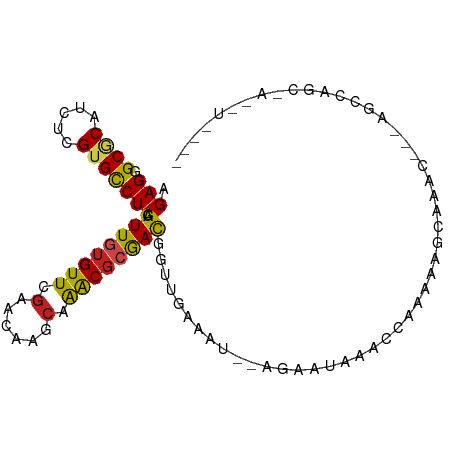
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.54 |
| Covariance contribution | 0.06 |
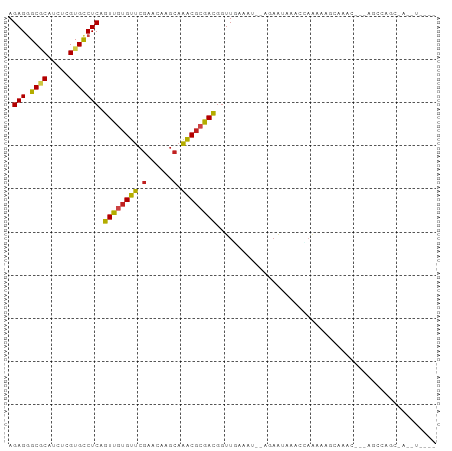
| Combinations/Pair | 1.38 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
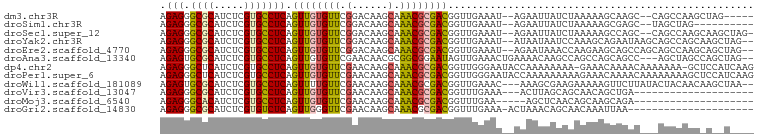
>dm3.chr3R 13947766 90 + 27905053 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGGACAAGCAAACGCGACGGUUGAAAU--AGAAUUAUCUAAAAAGCAAGC--CAGCCAAGCUAG----- .((((.(((.....))))))).((((((((.(......).))))))))(((((...(--(((....))))....(....)--))))).......----- ( -24.00, z-score = -0.68, R) >droSim1.chr3R 19981028 85 - 27517382 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGGACAAGCAAACGCGACGGUUGAAAU--AGAAUUAUCUAAAAAGCGAGC--UAGCUAG---------- .((((.(((.....)))))))(((((.((((((((..((....))....)))....(--(((....)))).....)))))--)))))..---------- ( -22.60, z-score = -0.35, R) >droSec1.super_12 1913887 94 - 2123299 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGGACAAGCAAACGCGACGGUUGAAAU--AGAAUUAUCUAAAAAGCCAGC--CAGCCAAGCAAGCUAG- .((((.(((.....))))))).((((((((.(......).))))))))((((....(--(((....))))...))))(((--..((...))..)))..- ( -26.20, z-score = -0.86, R) >droYak2.chr3R 2549188 95 + 28832112 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGGACAAGCAAACGCGACGGUUGAAAU--AUAAUAAUCCAAAGCAGAAUAAGCAGCCAGCAAGCUAG-- .((((.(((.....))))))).((((((((.(......).))))))))(((((...(--((................)))..)))))..........-- ( -22.29, z-score = 0.03, R) >droEre2.scaffold_4770 10058434 95 - 17746568 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGGACAAGCAAACGCGACGGUUGAAAU--AGAAUAAACCAAGAAGCAGCCAGCAGCCAAGCAGCUAG-- .((((.(((.....))))))).((((((((.(......).))))))))(((((....--................)))))(((.((...)).)))..-- ( -24.95, z-score = -0.08, R) >droAna3.scaffold_13340 11919718 94 - 23697760 AGAGUGCGCAUCUCGUGCCUCAGUUGUGUUCGAACAACGCGGCGGAAUAGUUGAAACUGAAAACAAGCCAGCCAGCAGCC---AGCUAGCCAGCUAG-- .(((.((((.....))))))).(((((......)))))(((((....((((....)))).......))).((.(((....---.))).))..))...-- ( -26.30, z-score = 0.20, R) >dp4.chr2 29032333 97 + 30794189 AGAGGGCUCAUCUCGUGCCUCAGUUGUGUUCGAACAAGCAAACGCGACGGUUGGGAAUACCAAAAAAAA-GAAACAAAACAAAAAAA-GCUCCAUCAAG .((((((.(.....).)))((.((((((((.(......).))))))))..((((.....))))......-))...............-.)))....... ( -18.80, z-score = -0.39, R) >droPer1.super_6 4357222 99 + 6141320 AGAGGGCUCAUCUCGUGCCUCAGUUGUGUUCGAACAAGCAAACGCGACGGUUGGGAAUACCAAAAAAAAAGAAACAAAACAAAAAAAAGCUCCAUCAAG .((((((.(.....).)))((.((((((((.(......).))))))))..((((.....)))).......)).................)))....... ( -18.80, z-score = -0.47, R) >droWil1.scaffold_181089 7606288 94 - 12369635 AGAGUGCGCAUCUCGUGCCUCAGUUUUGUUCGAACAAGCAAACGCGACGGUUGAAAC---AAAGCGAAGAAAAAGUUCUUAUACUACAACAAGCUAA-- .(((.((((.....))))))).((((((((..(((..((....))....)))..)))---))))).(((((....))))).................-- ( -22.60, z-score = -0.66, R) >droVir3.scaffold_13047 12508048 76 - 19223366 AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGAACAAGCAAACGCGACGGUUUGAAA---ACUUAGCAGCAACAGCUGA-------------------- ...((((((.....))))))(((((((.(((((((..((....))....))))))).---...........))))))).-------------------- ( -23.92, z-score = -0.73, R) >droMoj3.scaffold_6540 31583287 74 - 34148556 AGAGGGCACAUCUCGUGCCUCAGUUGUGUUCGAACAAGCAAACGCGACGGUUUGAA-----AGCUCAACAGCAAGCAGA-------------------- .((((.(((.....))))))).(((((.(((((((..((....))....)))))))-----......))))).......-------------------- ( -22.50, z-score = -1.00, R) >droGri2.scaffold_14830 3672897 77 - 6267026 AGAGGGCGCAUCUCGUGUCUCAGUUGGGUUCGAACAAGCAAACGCGACGGUUUGAAA-ACUAAACAGCAACAAAUUAA--------------------- .((((.(((.....))))))).((((..(((((((..((....))....))))))).-......))))..........--------------------- ( -17.70, z-score = -0.64, R) >consensus AGAGGGCGCAUCUCGUGCCUCAGUUGUGUUCGAACAAGCAAACGCGACGGUUGAAAU__AGAAUAAACCAAAAAGCAAAC___AGCCAGC_A__U____ .(((.((((.....))))))).((((((((.(......).))))))))................................................... (-15.48 = -15.54 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:32 2011