| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,937,347 – 13,937,402 |
| Length | 55 |
| Max. P | 0.998695 |

| Location | 13,937,347 – 13,937,402 |
|---|---|
| Length | 55 |
| Sequences | 8 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 65.87 |
| Shannon entropy | 0.64999 |
| G+C content | 0.43999 |
| Mean single sequence MFE | -18.26 |
| Consensus MFE | -7.24 |
| Energy contribution | -7.47 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
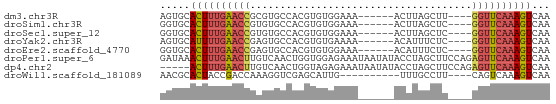
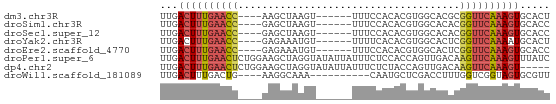
>dm3.chr3R 13937347 55 + 27905053 AGUGCACUUUGAACCGCGUGCCACGUGUGGAAA------ACUUAGCUU----GGUUCAAAGUCAA ..((.(((((((((((.((.(((....)))...------.....)).)----)))))))))))). ( -18.40, z-score = -2.15, R) >droSim1.chr3R 19969481 55 - 27517382 GGUGCACUUUGAACCGUGUGCCACGUGUGGAAA------ACUUAGCUC----GGUUCAAAGUCAA ..((.(((((((((((.((.(((....)))...------.....)).)----)))))))))))). ( -20.80, z-score = -3.05, R) >droSec1.super_12 1894454 55 - 2123299 GGUGCACUUUGAACCGUGUGCCACGUGUGGAAA------ACUUAGCUC----GGUUCAAAGUCAA ..((.(((((((((((.((.(((....)))...------.....)).)----)))))))))))). ( -20.80, z-score = -3.05, R) >droYak2.chr3R 2534138 55 + 28832112 AGUGCAUUUUGAACCGAGUGCCACGUGUGAAAA------ACAUUUCUC----GGUUCAAAGUCAA ..((.(((((((((((((......((((.....------))))..)))----)))))))))))). ( -20.00, z-score = -4.09, R) >droEre2.scaffold_4770 10048145 55 - 17746568 GGUGCACUUUGAACCGAGUGCCACGUGUGGAAA------ACAUUUCUC----GGUUCAAAGUCAA ..((.(((((((((((((..(((....)))...------......)))----)))))))))))). ( -22.60, z-score = -3.95, R) >droPer1.super_6 4339877 65 + 6141320 GAUAAACUUUGAACUUGUCAACUGGUGGAGAAAUAAUAUACCUAGCUUCCAGAGUUCAAAGUCAA .....(((((((((((.....((((((.(....)....)))).))......)))))))))))... ( -16.20, z-score = -2.50, R) >dp4.chr2 29017107 60 + 30794189 -----ACUUUGAACUUGUCAACUGGUAGAGAAAUAAUAUACCUAGCUUCCAGAGUUCAAAGUCAA -----(((((((((((.....((((((.(....)....)))).))......)))))))))))... ( -16.20, z-score = -2.98, R) >droWil1.scaffold_181089 7596001 51 - 12369635 AACGCACUACCGACCAAAGGUCGAGCAUUG----------UUUGCCUU----CAGUCAAAGUCAA ...........(((..((((.(((((...)----------))))))))----..)))........ ( -11.10, z-score = -1.48, R) >consensus GGUGCACUUUGAACCGAGUGCCACGUGUGGAAA______ACUUAGCUC____GGUUCAAAGUCAA .....((((((((((.....................................))))))))))... ( -7.24 = -7.47 + 0.22)
| Location | 13,937,347 – 13,937,402 |
|---|---|
| Length | 55 |
| Sequences | 8 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 65.87 |
| Shannon entropy | 0.64999 |
| G+C content | 0.43999 |
| Mean single sequence MFE | -18.25 |
| Consensus MFE | -6.93 |
| Energy contribution | -7.15 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.998695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
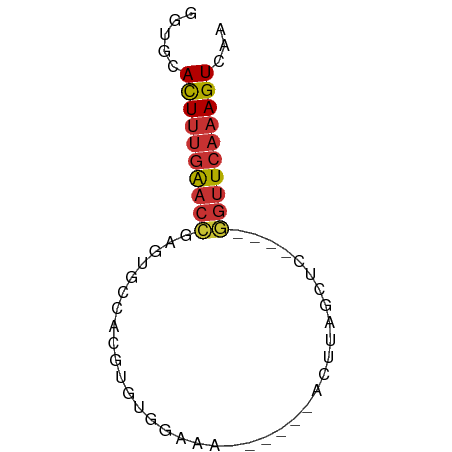
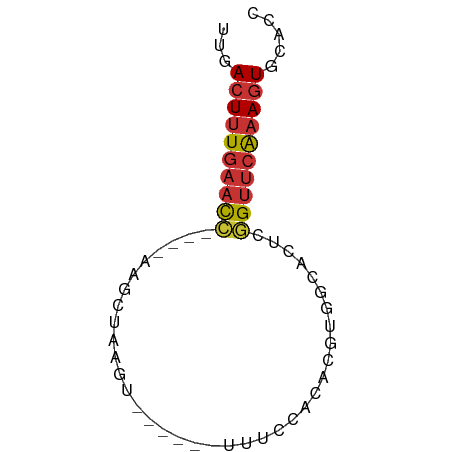
>dm3.chr3R 13937347 55 - 27905053 UUGACUUUGAACC----AAGCUAAGU------UUUCCACACGUGGCACGCGGUUCAAAGUGCACU .((((((((((((----..((((.((------.......)).))))....)))))))))).)).. ( -16.70, z-score = -2.11, R) >droSim1.chr3R 19969481 55 + 27517382 UUGACUUUGAACC----GAGCUAAGU------UUUCCACACGUGGCACACGGUUCAAAGUGCACC .((((((((((((----(.((((.((------.......)).))))...))))))))))).)).. ( -19.40, z-score = -3.32, R) >droSec1.super_12 1894454 55 + 2123299 UUGACUUUGAACC----GAGCUAAGU------UUUCCACACGUGGCACACGGUUCAAAGUGCACC .((((((((((((----(.((((.((------.......)).))))...))))))))))).)).. ( -19.40, z-score = -3.32, R) >droYak2.chr3R 2534138 55 - 28832112 UUGACUUUGAACC----GAGAAAUGU------UUUUCACACGUGGCACUCGGUUCAAAAUGCACU .....((((((((----(((..((((------.......))))....)))))))))))....... ( -16.60, z-score = -2.97, R) >droEre2.scaffold_4770 10048145 55 + 17746568 UUGACUUUGAACC----GAGAAAUGU------UUUCCACACGUGGCACUCGGUUCAAAGUGCACC .((((((((((((----(((..((((------.......))))....))))))))))))).)).. ( -21.40, z-score = -4.06, R) >droPer1.super_6 4339877 65 - 6141320 UUGACUUUGAACUCUGGAAGCUAGGUAUAUUAUUUCUCCACCAGUUGACAAGUUCAAAGUUUAUC ..(((((((((((.((..((((.(((.............)))))))..))))))))))))).... ( -19.52, z-score = -3.62, R) >dp4.chr2 29017107 60 - 30794189 UUGACUUUGAACUCUGGAAGCUAGGUAUAUUAUUUCUCUACCAGUUGACAAGUUCAAAGU----- ...((((((((((.((..((((.((((...........))))))))..))))))))))))----- ( -19.80, z-score = -3.83, R) >droWil1.scaffold_181089 7596001 51 + 12369635 UUGACUUUGACUG----AAGGCAAA----------CAAUGCUCGACCUUUGGUCGGUAGUGCGUU ...((((((((..----(((((...----------........).))))..))))).)))..... ( -13.20, z-score = -1.05, R) >consensus UUGACUUUGAACC____AAGCUAAGU______UUUCCACACGUGGCACUCGGUUCAAAGUGCACC ...((((((((((.....................................))))))))))..... ( -6.93 = -7.15 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:32 2011