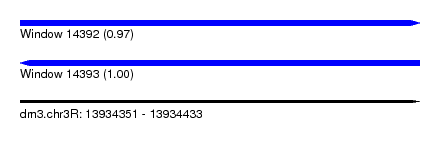
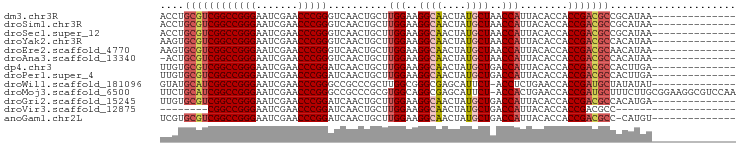
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,934,351 – 13,934,433 |
| Length | 82 |
| Max. P | 0.995474 |

| Location | 13,934,351 – 13,934,433 |
|---|---|
| Length | 82 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
| Shannon entropy | 0.34586 |
| G+C content | 0.57295 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
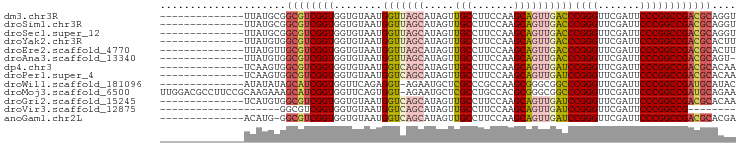
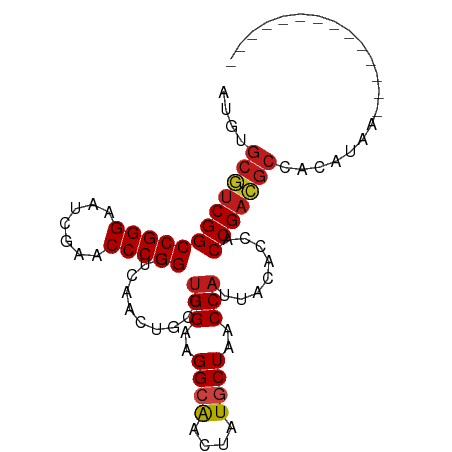
>dm3.chr3R 13934351 82 + 27905053 --------------UUAUGCGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCAGGU --------------.......((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).... ( -30.40, z-score = -1.26, R) >droSim1.chr3R 19965873 82 - 27517382 --------------UUAUGCGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCAGGU --------------.......((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).... ( -30.40, z-score = -1.26, R) >droSec1.super_12 1891408 82 - 2123299 --------------UUAUGCGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCAGGU --------------.......((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).... ( -30.40, z-score = -1.26, R) >droYak2.chr3R 2531224 82 + 28832112 --------------UUAUGUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCACUU --------------.......((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).... ( -30.40, z-score = -1.80, R) >droEre2.scaffold_4770 10045337 82 - 17746568 --------------UUAUGUUGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCACUU --------------......(((((((((........(((((((...(((........))).)))))))((((.......)))))))))))))... ( -31.00, z-score = -2.45, R) >droAna3.scaffold_13340 11907199 81 - 23697760 --------------UUAUGUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCAGU- --------------.......((((((((........(((((((...(((........))).)))))))((((.......))))))))))))...- ( -30.40, z-score = -1.98, R) >dp4.chr3 4489168 82 - 19779522 --------------UCAAGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAA --------------....(((.(((((((........(((((((...(((........))).)))))))((((.......)))))))))))))).. ( -30.70, z-score = -2.11, R) >droPer1.super_4 3761212 82 + 7162766 --------------UCAAGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAA --------------....(((.(((((((........(((((((...(((........))).)))))))((((.......)))))))))))))).. ( -30.70, z-score = -2.11, R) >droWil1.scaffold_181096 7895064 81 - 12416693 --------------AUAUAUAGCAUCGGUGGUUCAGAGGU-AGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAC --------------.......(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).... ( -38.10, z-score = -2.93, R) >droMoj3.scaffold_6500 9161665 95 - 32352404 UUGGACGCCUUCCGCAAGAAAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGAA ((((..(((.(((((..(((.((....))..))).(((((-((.........)))))))))))).)))(((((.......)))))))))....... ( -37.90, z-score = -0.25, R) >droGri2.scaffold_15245 14882096 82 - 18325388 --------------UCAUGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAA --------------...((((.(((((((........(((((((...(((........))).)))))))((((.......))))))))))))))). ( -31.50, z-score = -2.09, R) >droVir3.scaffold_12875 10314205 68 - 20611582 --------------------GGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCG-------- --------------------((((((((..((....(((...(((....)))...))).))..))))).((((.......))))))).-------- ( -20.90, z-score = -0.43, R) >anoGam1.chr2L 4571866 81 - 48795086 --------------ACAUG-GGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACGA --------------.....-.((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).... ( -30.00, z-score = -1.00, R) >consensus ______________UUAUGUGGCGUCGGUGGUGUAAUGGUUAGCAUAGUUGCCUUCCAAGCAGUUGACCCGGGUUCGAUUCCCGGCCGACGCACAA .....................((((((((........(((((((.....(((.......))))))))))((((.......)))))))))))).... (-28.38 = -28.26 + -0.12)
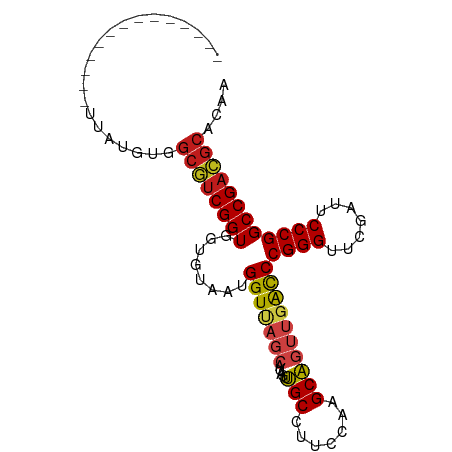
| Location | 13,934,351 – 13,934,433 |
|---|---|
| Length | 82 |
| Sequences | 13 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.34 |
| Shannon entropy | 0.34586 |
| G+C content | 0.57295 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13934351 82 - 27905053 ACCUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCGCAUAA-------------- ....((((((((((((.......)))))..........(((..((((....))))..)))........))))))).......-------------- ( -28.10, z-score = -2.11, R) >droSim1.chr3R 19965873 82 + 27517382 ACCUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCGCAUAA-------------- ....((((((((((((.......)))))..........(((..((((....))))..)))........))))))).......-------------- ( -28.10, z-score = -2.11, R) >droSec1.super_12 1891408 82 + 2123299 ACCUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCGCAUAA-------------- ....((((((((((((.......)))))..........(((..((((....))))..)))........))))))).......-------------- ( -28.10, z-score = -2.11, R) >droYak2.chr3R 2531224 82 - 28832112 AAGUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCACAUAA-------------- ..((((((((((((((.......)))))..........(((..((((....))))..)))........)))))).)))....-------------- ( -29.70, z-score = -2.85, R) >droEre2.scaffold_4770 10045337 82 + 17746568 AAGUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCAACAUAA-------------- ...(((((((((((((.......)))))..........(((..((((....))))..)))........))))))))......-------------- ( -29.90, z-score = -3.16, R) >droAna3.scaffold_13340 11907199 81 + 23697760 -ACUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCACAUAA-------------- -...((((((((((((.......)))))..........(((..((((....))))..)))........))))))).......-------------- ( -28.10, z-score = -2.85, R) >dp4.chr3 4489168 82 + 19779522 UUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACUUGA-------------- .((.((((((((((((.......)))))..........(((..((((....))))..)))........))))))))).....-------------- ( -29.90, z-score = -3.29, R) >droPer1.super_4 3761212 82 - 7162766 UUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACUUGA-------------- .((.((((((((((((.......)))))..........(((..((((....))))..)))........))))))))).....-------------- ( -29.90, z-score = -3.29, R) >droWil1.scaffold_181096 7895064 81 + 12416693 GUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCU-ACCUCUGAACCACCGAUGCUAUAUAU-------------- ((((((((((((((((.......)))))(((((((((...))))))).)).....-............))))))).))))..-------------- ( -39.40, z-score = -3.97, R) >droMoj3.scaffold_6500 9161665 95 + 32352404 UUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUUUCUUGCGGAAGGCGUCCAA .......(((((((((.......)))))((((((.((...)).)))).)).....-....))))......(((((((((.....)))))))))... ( -34.70, z-score = -0.17, R) >droGri2.scaffold_15245 14882096 82 + 18325388 UUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACAUGA-------------- .(((((((((((((((.......)))))..........(((..((((....))))..)))........)))))).))))...-------------- ( -31.00, z-score = -3.53, R) >droVir3.scaffold_12875 10314205 68 + 20611582 --------CGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCC-------------------- --------((((((((.......)))))..........(((..((((....))))..)))........))).....-------------------- ( -18.80, z-score = -1.43, R) >anoGam1.chr2L 4571866 81 + 48795086 UCGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCC-CAUGU-------------- ..(.((((((((((((.......)))))..........(((..((((....))))..)))........))))))).-)....-------------- ( -29.60, z-score = -2.43, R) >consensus AUGUGCGUCGGCCGGGAAUCGAACCCGGGUCAACUGCUUGGAAGGCAACUAUGCUAACCAUUACACCACCGACGCCACAUAA______________ ....((((((((((((.......)))))..........(((..((((....))))..)))........)))))))..................... (-25.90 = -26.07 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:29 2011