
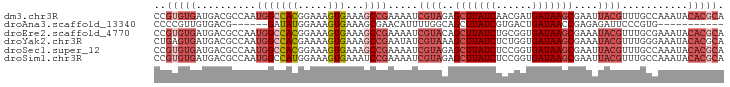
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,920,211 – 13,920,306 |
| Length | 95 |
| Max. P | 0.879874 |

| Location | 13,920,211 – 13,920,306 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Shannon entropy | 0.42504 |
| G+C content | 0.47067 |
| Mean single sequence MFE | -25.40 |
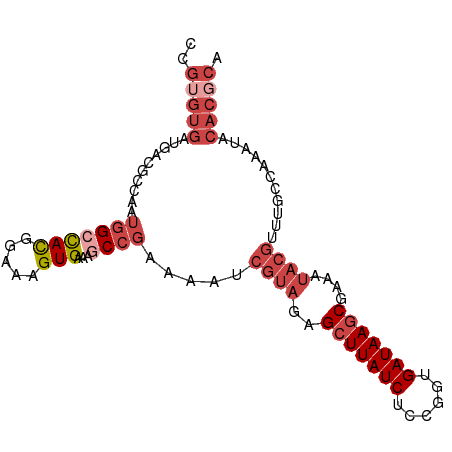
| Consensus MFE | -14.84 |
| Energy contribution | -16.98 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
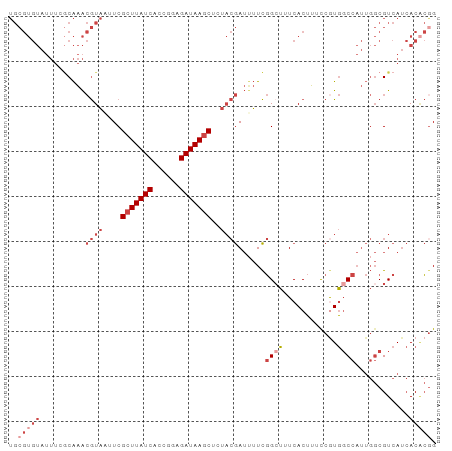
>dm3.chr3R 13920211 95 + 27905053 CCGUGUGAUGACGCCAAUGGCCACGGAAAGUGAAAGCCGAAAAUCGUAGAGCUUAUCAACGAUGAUAAGCGAAUUACGUUUGCCAAAUACACGCA ..(((((((...((...(((((((.....)))...)))).....(((((.((((((((....))))))))...)))))...))...)).))))). ( -27.20, z-score = -2.62, R) >droAna3.scaffold_13340 11893226 78 - 23697760 CCCCGUUGUGACG------GAUAUGGAAAGUGAAAGCGAACAUUUUGGCAGCUUAUCGUGACUGAUAACCGAGAGAUUCCCGUG----------- ..((((....)))------).(((((...((....))(((..((((((....((((((....))))))))))))..))))))))----------- ( -18.20, z-score = -1.11, R) >droEre2.scaffold_4770 10031389 95 - 17746568 CCGUGUGAUGACGCCAAUGGCCACGGAAAGUGAAAGCCGAAAAUCGUACAGCUUAUCUGCGGUGAUAAGCGAAAUACGUUUGCGAAAUACACGCA ..(((((((..(((...(((((((.....)))...)))).....((((..(((((((......)))))))....))))...)))..)).))))). ( -29.00, z-score = -1.93, R) >droYak2.chr3R 2516964 95 + 28832112 CUGAGUGAUGACGCCAAUGGCCACGAAAAGUGAAAGCCGAAUAUCGUAAAGCUUAUCUCUGGUGAUAAGCGAAAUACGUUUGGGAAAUACACGCA ....(((((..(.(((((((((((.....)))...)))).....((((..(((((((......)))))))....)))).)))))..)).)))... ( -24.50, z-score = -1.72, R) >droSec1.super_12 1877270 95 - 2123299 CCGUGUGAUGACGCCAAUGGCCACGGAAAGUGAAAGCCGAAAAUCGUAGAGCUUAUCUCCGGUGAUAAGCGAAUUACGUUUGCCAAAUACACGCA ..(((((((...((...(((((((.....)))...)))).....(((((.(((((((......)))))))...)))))...))...)).))))). ( -27.00, z-score = -1.75, R) >droSim1.chr3R 19951853 95 - 27517382 CCGUGUGAUGACGCCAAUGGCCAUGGAAAGUGAAAUCCGAAAAUCGUAGAGCUUAUCUCCGGUGAUAAGCGAAUUACGUUUGCCAAAUACACGCA ..(((((.((....)).((((..((((........)))).....(((((.(((((((......)))))))...)))))...))))....))))). ( -26.50, z-score = -1.92, R) >consensus CCGUGUGAUGACGCCAAUGGCCACGGAAAGUGAAAGCCGAAAAUCGUAGAGCUUAUCUCCGGUGAUAAGCGAAAUACGUUUGCCAAAUACACGCA ..(((((..........(((((((.....)))...)))).....((((..(((((((......)))))))....))))...........))))). (-14.84 = -16.98 + 2.14)

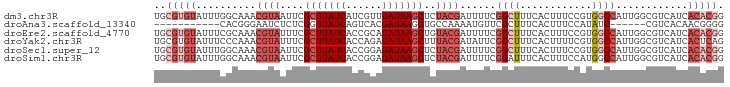
| Location | 13,920,211 – 13,920,306 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Shannon entropy | 0.42504 |
| G+C content | 0.47067 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -11.59 |
| Energy contribution | -13.95 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13920211 95 - 27905053 UGCGUGUAUUUGGCAAACGUAAUUCGCUUAUCAUCGUUGAUAAGCUCUACGAUUUUCGGCUUUCACUUUCCGUGGCCAUUGGCGUCAUCACACGG ..(((((...((((...((((....((((((((....))))))))..))))......(((...(((.....))))))......))))..))))). ( -29.10, z-score = -3.16, R) >droAna3.scaffold_13340 11893226 78 + 23697760 -----------CACGGGAAUCUCUCGGUUAUCAGUCACGAUAAGCUGCCAAAAUGUUCGCUUUCACUUUCCAUAUC------CGUCACAACGGGG -----------...(((((......(.(((((......))))).).((.((....)).))......)))))...((------(((....))))). ( -12.60, z-score = 0.37, R) >droEre2.scaffold_4770 10031389 95 + 17746568 UGCGUGUAUUUCGCAAACGUAUUUCGCUUAUCACCGCAGAUAAGCUGUACGAUUUUCGGCUUUCACUUUCCGUGGCCAUUGGCGUCAUCACACGG ..(((((((..(((...(((((...(((((((......))))))).)))))......(((...(((.....))))))....)))..)).))))). ( -29.90, z-score = -3.02, R) >droYak2.chr3R 2516964 95 - 28832112 UGCGUGUAUUUCCCAAACGUAUUUCGCUUAUCACCAGAGAUAAGCUUUACGAUAUUCGGCUUUCACUUUUCGUGGCCAUUGGCGUCAUCACUCAG ((.(((.((..(((((.((((....(((((((......)))))))..))))......(((...(((.....)))))).)))).)..))))).)). ( -24.50, z-score = -2.91, R) >droSec1.super_12 1877270 95 + 2123299 UGCGUGUAUUUGGCAAACGUAAUUCGCUUAUCACCGGAGAUAAGCUCUACGAUUUUCGGCUUUCACUUUCCGUGGCCAUUGGCGUCAUCACACGG ..(((((...((((...((((....(((((((......)))))))..))))......(((...(((.....))))))......))))..))))). ( -27.60, z-score = -2.09, R) >droSim1.chr3R 19951853 95 + 27517382 UGCGUGUAUUUGGCAAACGUAAUUCGCUUAUCACCGGAGAUAAGCUCUACGAUUUUCGGAUUUCACUUUCCAUGGCCAUUGGCGUCAUCACACGG ..(((((...((((...((((....(((((((......)))))))..))))......(((........)))...(((...)))))))..))))). ( -27.50, z-score = -2.53, R) >consensus UGCGUGUAUUUCGCAAACGUAAUUCGCUUAUCACCGGAGAUAAGCUCUACGAUUUUCGGCUUUCACUUUCCGUGGCCAUUGGCGUCAUCACACGG ..(((((..........((((....(((((((......)))))))..))))......((((............))))............))))). (-11.59 = -13.95 + 2.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:26 2011