
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,854,460 – 7,854,555 |
| Length | 95 |
| Max. P | 0.533540 |

| Location | 7,854,460 – 7,854,555 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.75 |
| Shannon entropy | 0.54457 |
| G+C content | 0.37486 |
| Mean single sequence MFE | -16.51 |
| Consensus MFE | -10.09 |
| Energy contribution | -9.90 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
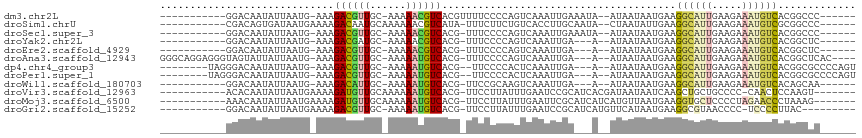
>dm3.chr2L 7854460 95 - 23011544 -----------GGACAAUAUUAAUG-AAAGACGUUGC-AAAAACGUCACGUUUUCCCCAGUCAAAUUGAAAUA--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCCC------ -----------((.((.(((((...-...((((((..-...))))))..........((((...)))).....--.))))).))..((((((.....))))))....)).------ ( -16.30, z-score = -0.57, R) >droSim1.chrU 13216895 96 - 15797150 -----------CGACAGUGAUAAUGAAAAGACAAUGCAAAAAACGUCAUA-UUUCUUCUGUCACCUUGCAAUA--CUAAUAUUGAAGGCAUUGAAGAAAUGUCGCGGCCC------ -----------................................(((.(((-(((((((.....((((.(((((--....)))))))))....)))))))))).)))....------ ( -20.70, z-score = -1.63, R) >droSec1.super_3 3368825 94 - 7220098 -----------GGACAAUAUUAAUG-AAAGACGUUGC-AAAAACGUCACG-UUUCCCCAGUCAAAUUGAAAUA--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCCC------ -----------((.((.(((((...-...((((((..-...))))))..(-((((............))))).--.))))).))..((((((.....))))))....)).------ ( -17.20, z-score = -1.01, R) >droYak2.chr2L 17282184 91 + 22324452 -----------GGACAAUAUUAAUG-AAAGACGAUGC-AAAAACGUCACG-UUUCCCCAGUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCUC------ -----------.(((......((((-...((((....-.....)))).))-))......))).......---.--...........((((((.....)))))).......------ ( -12.10, z-score = 0.73, R) >droEre2.scaffold_4929 16776065 91 - 26641161 -----------GGACAAUAUUAAUG-AAAGACGUUGC-AAAAACGUCACG-UUUCCCCAGUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCUC------ -----------.(((......((((-...((((((..-...)))))).))-))......))).......---.--...........((((((.....)))))).......------ ( -16.30, z-score = -0.60, R) >droAna3.scaffold_12943 3589303 104 + 5039921 GGGCAGGAGGGUAGUAUUAUUAAUG-AAAGACGUUGC-AAAAAUGUCACG-UUUCCCCAGUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCUCAC---- ((((.((.(((..((.(((....))-)..((((((..-...)))))))).-..)))))...........---.--...........((((((.....))))))...))))..---- ( -21.60, z-score = -0.73, R) >dp4.chr4_group3 6249878 99 + 11692001 --------UAGGGACAAUAUUAAUG-AAAGACGUUGC-AAAAAUGUCACG--UUCCCCACUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCGCCCCAGU --------..(((.((.(((((...-...((((((..-...))))))...--........((.....))---.--.))))).))..((((((.....)))))).......)))... ( -18.70, z-score = -0.51, R) >droPer1.super_1 3350106 99 + 10282868 --------UAGGGACAAUAUUAAUG-AAAGACGUUGC-AAAAAUGUCACG--UUCCCCACUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCGCCCCAGU --------..(((.((.(((((...-...((((((..-...))))))...--........((.....))---.--.))))).))..((((((.....)))))).......)))... ( -18.70, z-score = -0.51, R) >droWil1.scaffold_180703 466594 91 + 3946847 -----------GGACAAUAUUAAUG-AAAGACAUUGC-AAAAAUGUCACG-UUCCGCAAGUCAAAUUGA---A--AUAAUAAUGAAGGCAUUGAAGAAAUGUCACAGCAA------ -----------...((.(((((...-...((((((..-...))))))...-...(....).........---.--.))))).))..((((((.....)))))).......------ ( -15.30, z-score = -0.81, R) >droVir3.scaffold_12963 8619592 96 - 20206255 -----------ACACAAUAUUAAUGAAAAGAUGUUGCAAAAAAUGUCACG-UUCCUUAUUUGAAUCCGCAUCACGAUAAUAAUCAAGCUGCUGCCCC-CAACUCCAAGU------- -----------...(((...(((.(((..((((((......))))))...-))).))).))).....(((.(..(((....)))..).)))......-...........------- ( -8.90, z-score = 0.76, R) >droMoj3.scaffold_6500 29628055 97 + 32352404 -----------AAACAAUAUUAAUGAAAAGAUGUUGCAAAAAAUGUCACG-UUCCUUAUUUGAAUUCGCAUCAUCAUGUUAAUGAAGGUGCUCCCCUAGAACCCUAAAG------- -----------...(((...(((.(((..((((((......))))))...-))).))).))).....(((((.((((....)))).)))))..................------- ( -16.40, z-score = -1.90, R) >droGri2.scaffold_15252 4017291 93 + 17193109 -----------GGACAAUAUUAAUGAAAAGACGUUGC-AAAAAUGUCACG-UUCCUUAUUUGAAUCCGCAUCAUGUUCAUAAUGAAGGCGUAACCCC-UCCCCUUAC--------- -----------((((((...(((.(((..((((((..-...))))))...-))).))).)))....(((.((((.......))))..))).......-)))......--------- ( -15.90, z-score = -0.94, R) >consensus ___________GGACAAUAUUAAUG_AAAGACGUUGC_AAAAAUGUCACG_UUUCCCCAGUCAAAUUGA___A__AUAAUAAUGAAGGCAUUGAAGAAAUGUCACGGCUC______ .............................((((((......)))))).......................................((((((.....))))))............. (-10.09 = -9.90 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:32 2011