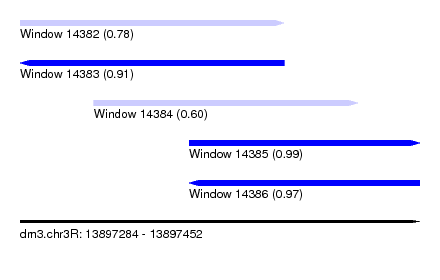
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,897,284 – 13,897,452 |
| Length | 168 |
| Max. P | 0.989625 |

| Location | 13,897,284 – 13,897,395 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.11 |
| Shannon entropy | 0.23380 |
| G+C content | 0.37693 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.19 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13897284 111 + 27905053 --------CUCGGCUGAGACAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAAUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------.(((......((((((....((((((((((.......))))))).)))...((....))....)))))).......(((((((((((....)).)))))))))....))). ( -28.80, z-score = -1.88, R) >droSim1.chr3R 19923101 111 - 27517382 --------CUCGGCUGAGACAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------.(((......((((((....((((((((((.......))))))).))).((((....))))..)))))).......(((((((((((....)).)))))))))....))). ( -30.00, z-score = -2.23, R) >droSec1.super_12 1854503 111 - 2123299 --------CUCGGCUGAGACAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------.(((......((((((....((((((((((.......))))))).))).((((....))))..)))))).......(((((((((((....)).)))))))))....))). ( -30.00, z-score = -2.23, R) >droYak2.chr3R 2490852 111 + 28832112 --------CUCGGCUGAGACAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------.(((......((((((....((((((((((.......))))))).))).((((....))))..)))))).......(((((((((((....)).)))))))))....))). ( -30.00, z-score = -2.49, R) >droEre2.scaffold_4770 10008158 111 - 17746568 --------CUCGGCUGAGACAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------.(((......((((((....((((((((((.......))))))).))).((((....))))..)))))).......(((((((((((....)).)))))))))....))). ( -30.00, z-score = -2.49, R) >droAna3.scaffold_13340 11871716 111 - 23697760 --------CUCGGCUGGGGCAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGCCCCGCUGUGAAUUUGUGACAUUUUGGUAAACGAAAAAUGUCAGUUACGAU --------(.((((.(((((........((((((((((.......))))))).)))...((....))))))))))).)......(((((((((.........)))))))))........ ( -32.60, z-score = -2.27, R) >dp4.chr2 28957470 117 + 30794189 CCCGUCUUUUGGGCCGCAGCAU--UUCCAAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU ((((.....)))).((((((..--...(((.(((((((.......))))))).))).((((....))))...))))))......(((((((((((....)).)))))))))........ ( -33.90, z-score = -3.46, R) >droPer1.super_6 4282267 117 + 6141320 CCCGUCUUUUGGGCCGCAGCAU--UUCCAAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU ((((.....)))).((((((..--...(((.(((((((.......))))))).))).((((....))))...))))))......(((((((((((....)).)))))))))........ ( -33.90, z-score = -3.46, R) >droWil1.scaffold_181089 7563234 110 - 12369635 --------CUCGCCCG-AUCGUUCGUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAAUCCCGCUGUGAAUUUGUGACAUUUUUGUAAACGAAAAAUGUCAGUUACUAU --------...(((((-(....)))...((((((((((.......))))))).)))....)))............((((.....((((((((((.......)))))))))).))))... ( -20.90, z-score = -1.09, R) >droVir3.scaffold_13047 12441040 107 - 19223366 --------CUGGACCGGGGCAU--UUC--AAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU --------......((((((..--..(--(((((((((.......))))))).)))...((....)))))))).(((((.....(((((((((((....)).))))))))).))))).. ( -29.70, z-score = -2.54, R) >droMoj3.scaffold_6540 31513113 98 - 34148556 ------------------GCAU--UUCC-AAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU ------------------((..--...(-(((((((((.......))))))).))).((((....))))...))(((((.....(((((((((((....)).))))))))).))))).. ( -22.20, z-score = -1.93, R) >droGri2.scaffold_14830 3610319 107 - 6267026 --------UUCGGCUGGGGCA----UUUCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUGGUAAACGAAAAAUGUCAGUUACGAU --------..((((.(((((.----...((((((((((.......))))))).)))...((....)))))))))))........(((((((((.........)))))))))........ ( -29.00, z-score = -2.23, R) >consensus ________CUCGGCUGAGGCAGCGUUUCCAAUGAUUUAUUGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAU ...............................(((((((.......))))))).(((((((((((((((.....))))............((((((....)))))).)))))).))))). (-18.84 = -19.19 + 0.35)

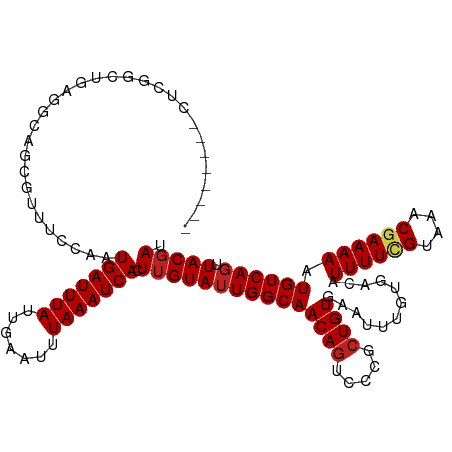
| Location | 13,897,284 – 13,897,395 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Shannon entropy | 0.23380 |
| G+C content | 0.37693 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13897284 111 - 27905053 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAACUCACAGCGGGACUGUUGCCAUUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGUCUCAGCCGAG-------- .(((....(((((((((.((....)))))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -28.00, z-score = -2.60, R) >droSim1.chr3R 19923101 111 + 27517382 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAACUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGUCUCAGCCGAG-------- .(((....(((((((((.((....)))))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -28.00, z-score = -2.64, R) >droSec1.super_12 1854503 111 + 2123299 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAACUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGUCUCAGCCGAG-------- .(((....(((((((((.((....)))))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -28.00, z-score = -2.64, R) >droYak2.chr3R 2490852 111 - 28832112 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGUCUCAGCCGAG-------- .(((....(((((((((.((....)))))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -28.00, z-score = -2.64, R) >droEre2.scaffold_4770 10008158 111 + 17746568 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGUCUCAGCCGAG-------- .(((....(((((((((.((....)))))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -28.00, z-score = -2.64, R) >droAna3.scaffold_13340 11871716 111 + 23697760 AUCGUAACUGACAUUUUUCGUUUACCAAAAUGUCACAAAUUCACAGCGGGGCUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGCCCCAGCCGAG-------- .(((....(((((((((.........)))))))))..........(((((((....((.......(((((((((.......)))))))))(....))).))))).))))).-------- ( -29.50, z-score = -2.92, R) >dp4.chr2 28957470 117 - 30794189 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUUGGAA--AUGCUGCGGCCCAAAAGACGGG .((((...(((((((((.((....)))))))))))............(((.((((.((.....(((((((((((.......)))))))))))...--..)).))))))).....)))). ( -35.90, z-score = -4.25, R) >droPer1.super_6 4282267 117 - 6141320 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUUGGAA--AUGCUGCGGCCCAAAAGACGGG .((((...(((((((((.((....)))))))))))............(((.((((.((.....(((((((((((.......)))))))))))...--..)).))))))).....)))). ( -35.90, z-score = -4.25, R) >droWil1.scaffold_181089 7563234 110 + 12369635 AUAGUAACUGACAUUUUUCGUUUACAAAAAUGUCACAAAUUCACAGCGGGAUUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAACGAACGAU-CGGGCGAG-------- ........((((((((((.(....)))))))))))..........((..(((((((.(((......((((((((.......)))))))))))....))))))-)..))...-------- ( -23.60, z-score = -1.67, R) >droVir3.scaffold_13047 12441040 107 + 19223366 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUU--GAA--AUGCCCCGGUCCAG-------- ........(((((((((.((....))))))))))).............((((((..((.......(((((((((.......)))))))))--...--..))..))))))..-------- ( -26.60, z-score = -2.90, R) >droMoj3.scaffold_6540 31513113 98 + 34148556 AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUU-GGAA--AUGC------------------ .(((((..(((((((((.((....)))))))))))........(((((....)))))....))).(((((((((.......)))))))))-.)).--....------------------ ( -20.20, z-score = -1.45, R) >droGri2.scaffold_14830 3610319 107 + 6267026 AUCGUAACUGACAUUUUUCGUUUACCAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGAAA----UGCCCCAGCCGAA-------- .(((....(((((((((.........)))))))))..........(((((..(((.......)))(((((((((.......)))))))))....----...))).))))).-------- ( -19.70, z-score = -1.02, R) >consensus AUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAUUCACAGCGGGACUGUUGCCAAUACAAGUGAUUUAAAUUCAAUAAAUCAUUGGAAACGCUGCCUCAGCCGAG________ ...(((..((((((((((.......))))))))))........(((((....)))))....))).(((((((((.......)))))))))............................. (-17.60 = -17.71 + 0.10)

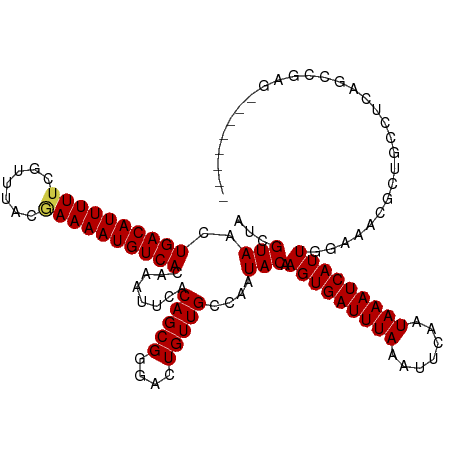
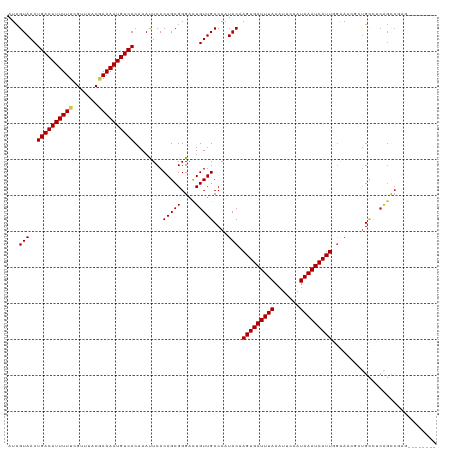
| Location | 13,897,315 – 13,897,426 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Shannon entropy | 0.35405 |
| G+C content | 0.37428 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13897315 111 + 27905053 UGAAUUUAAAUCACUUGUAAUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACCACAAACUCGGAUGUGC-GCU------- ......................(((((((.....))))((((((((((((((((((....)).)))))))).....((((.....))))...)))))))).....)))-...------- ( -29.20, z-score = -2.00, R) >droSim1.chr3R 19923132 112 - 27517382 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACCACAAACUCGGAUGUGCUGGU------- ..................((..(((((((.....))))((((((((((((((((((....)).)))))))).....((((.....))))...)))))))).....)))..))------- ( -32.10, z-score = -2.66, R) >droSec1.super_12 1854534 112 - 2123299 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAGUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACCACAAACUCGGAUGUGCUGGU------- ..................((..(((((((.....))))((((((((((((((((((....)).)))))))).....((((.....))))...)))))))).....)))..))------- ( -32.10, z-score = -2.66, R) >droYak2.chr3R 2490883 112 + 28832112 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACUACAAAUUCGAAUGUGCUGGU------- ..................((..(((((((.....))))((((((((((((((((((....)).)))))))).....((((.....))))...)))))))).....)))..))------- ( -29.70, z-score = -2.71, R) >droEre2.scaffold_4770 10008189 112 - 17746568 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACUACAAACUCGGAUGUGCUGGU------- ..................((..(((((((.....))))((.(((((((((((((((....)).)))))))).....((((.....))))...))))).)).....)))..))------- ( -26.00, z-score = -1.21, R) >droAna3.scaffold_13340 11871747 104 - 23697760 UGAAUUUAAAUCACUUGUAUUGGCAACAGCCCCGCUGUGAAUUUGUGACAUUUUGGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUGACUGCAGGCCCGAAG--------------- .............((((((.((((.(((((...))))).......(((((((((.........)))))))))))))((((.....))))..)))))).......--------------- ( -19.60, z-score = 0.08, R) >dp4.chr2 28957507 107 + 30794189 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUGACUUACAGCAGAGCCAAG------------ .............((((.((((....))))...(((((((.....(((((((((((....)).)))))))))....((((.....))))..))))))).....))))------------ ( -26.00, z-score = -2.36, R) >droPer1.super_6 4282304 107 + 6141320 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUGACUUACAGCAGAGCCAAA------------ ...................(((((.....((..(((((((.....(((((((((((....)).)))))))))....((((.....))))..))))))).))))))).------------ ( -25.40, z-score = -2.14, R) >droWil1.scaffold_181089 7563264 106 - 12369635 UGAAUUUAAAUCACUUGUAUUGGCAACAAUCCCGCUGUGAAUUUGUGACAUUUUUGUAAACGAAAAAUGUCAGUUACUAUUUUUUAUUGGCCCGAAGAAAAUGCAG------------- ..........((((..((((((....))))...)).)))).....((((((((((.......)))))))))).....((((((((.(((...)))))))))))...------------- ( -17.50, z-score = -0.01, R) >droVir3.scaffold_13047 12441067 107 - 19223366 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCU---GCACUCCCCAGUGCC--------- ................(((((((....(((.(.((((((((.((((((((((((((....)).))))))))....))))...)))).)))).---).)))..))))))).--------- ( -24.00, z-score = -1.98, R) >droMoj3.scaffold_6540 31513131 119 - 34148556 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCCCCAGCGCUGCCCAGCGUCGAGCCGAGC ...................(((((.........((((((((.((((((((((((((....)).))))))))....))))...)))).))))..(.(((((....))))).).))))).. ( -27.90, z-score = -1.18, R) >droGri2.scaffold_14830 3610346 114 - 6267026 UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUGGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCU---GCACUCCCCAGAGCCGA--CAAAC ..............((((...(((.((((.....))))...((((.((((((((.........))))))))(((((...........)))))---........))))))).)--))).. ( -19.50, z-score = 0.45, R) >consensus UGAAUUUAAAUCACUUGUAUUGGCAACAGUCCCGCUGUGAAUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUGACUACAAACUCGGACAUGC_GG________ ..........((((..((((((....))))...)).)))).....(((((((((((....)).)))))))))............................................... (-16.74 = -16.94 + 0.20)

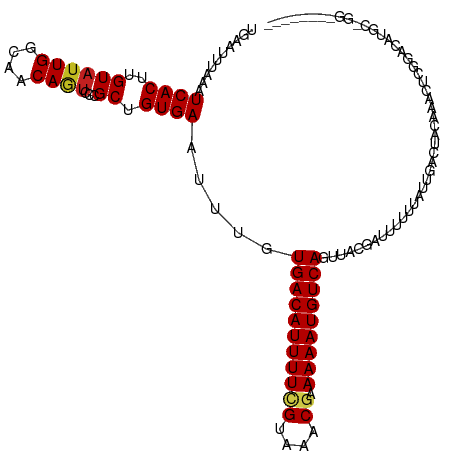
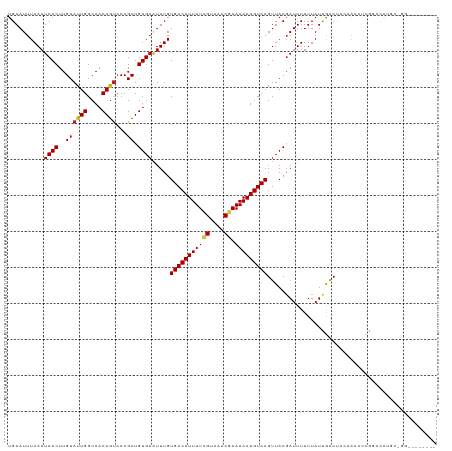
| Location | 13,897,355 – 13,897,452 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 61.22 |
| Shannon entropy | 0.74633 |
| G+C content | 0.40228 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -9.61 |
| Energy contribution | -9.63 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
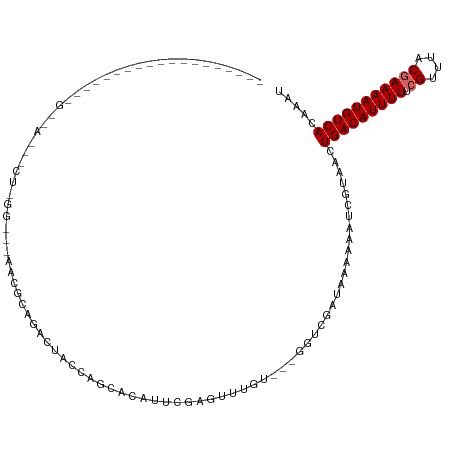
>dm3.chr3R 13897355 97 + 27905053 GUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACC---ACAAACUCGGAUGUGC-GCUGGUAUCCGUUCUGUCCAAAGUUUCCC------------------ ((((((((((((((((....)).)))))))).....((((.....))))...---))))))..((((..((-(........)))...))))..........------------------ ( -23.70, z-score = -1.80, R) >droSim1.chr3R 19923172 98 - 27517382 GUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACC---ACAAACUCGGAUGUGCUGGUAGUAUCCGUUCUGCCCAGAGUUUCCC------------------ ((((((((((((((((....)).)))))))).....((((.....))))...---))))))..(((....(((((((........))).))))....))).------------------ ( -28.40, z-score = -3.30, R) >droSec1.super_12 1854574 98 - 2123299 GUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACC---ACAAACUCGGAUGUGCUGGUAGUAUCCGUUCUGCCCAGAGUUUCCC------------------ ((((((((((((((((....)).)))))))).....((((.....))))...---))))))..(((....(((((((........))).))))....))).------------------ ( -28.40, z-score = -3.30, R) >droYak2.chr3R 2490923 83 + 28832112 AUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACU---ACAAAUUCGAAUGUGCUGGUAUUUUUUCCAA--------------------------------- ((((((((((((((((....)).)))))))).....((((.....))))...---))))))..........(((........))).--------------------------------- ( -16.50, z-score = -1.37, R) >droEre2.scaffold_4770 10008229 80 - 17746568 AUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACU---ACAAACUCGGAUGUGCUGGUCUUCUUCC------------------------------------ .....(((((((((((....)).))))))))).....(((.....)))((((---(...((......))..))))).......------------------------------------ ( -16.50, z-score = -1.23, R) >droWil1.scaffold_181089 7563304 80 - 12369635 AUUUGUGACAUUUUUGUAAACGAAAAAUGUCAGUUACUAUUUUUUAUUGGCC---CGAAGA--AAAUGCAGGGCCAGCGCGCAUC---------------------------------- .....((((((((((.......))))))))))..............((((((---(.....--.......)))))))........---------------------------------- ( -20.60, z-score = -1.80, R) >droVir3.scaffold_13047 12441107 96 - 19223366 AUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCU---GCACUCCCCAGUGCCG-----------------CCGUGGAGGCACU---GAUGAAGUUCAAGUU .....(((((((((((....)).))))))))).....((..(((((((((.(---((.((((.(.(.....-----------------).).)))))))))---))))))).))..... ( -28.30, z-score = -2.95, R) >droMoj3.scaffold_6540 31513171 116 - 34148556 AUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCCCCAGCGCUGCCCAGCGUCGAGCCGAGCCGCGUCGAGUCGAGCCGUCGCU---GAUGAAGUUCACGUU .....(((((((((((....)).))))))))).....((..((((((((((..(.(((((....))).((((..(((......)))..)))))).)..)))---))))))).))..... ( -34.10, z-score = -1.83, R) >droGri2.scaffold_14830 3610386 114 - 6267026 AUUUGUGACAUUUUGGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUUAGCU---GCACUCCCCAGAGCCGA--CAAACUGAUGAAGUCCAAGCUGCAGUUCGAGCUGAAGUUGAAGUU .....(((((((((.........)))))))))....(((((((...((((((---((((((....)))..((--(...........))).....))))))).))...)))))))..... ( -23.20, z-score = 0.10, R) >consensus AUUUGUGACAUUUUCGUAAACGAAAAAUGUCAGUUACGAUUUUUUAUCGACC___ACAAACUCGAAUGUGCUGGUAGUAUCCGUU___CC_AG___U__C___________________ .....(((((((((((....)).)))))))))....................................................................................... ( -9.61 = -9.63 + 0.03)

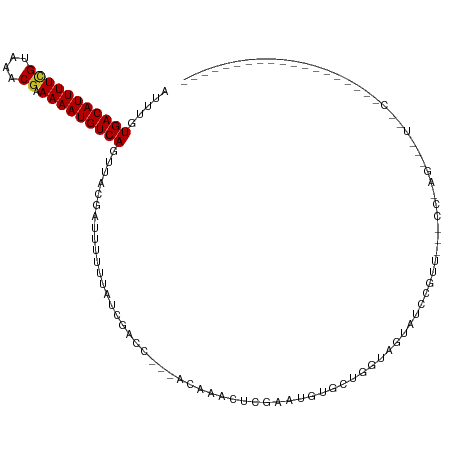
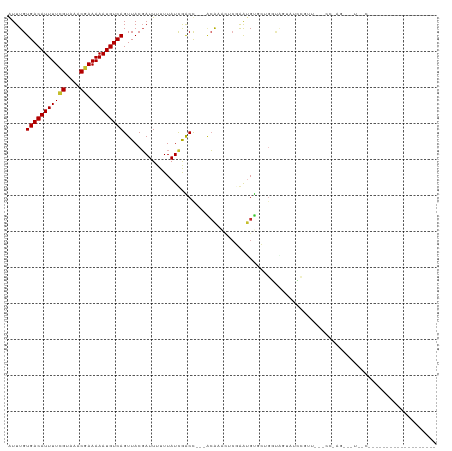
| Location | 13,897,355 – 13,897,452 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 61.22 |
| Shannon entropy | 0.74633 |
| G+C content | 0.40228 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
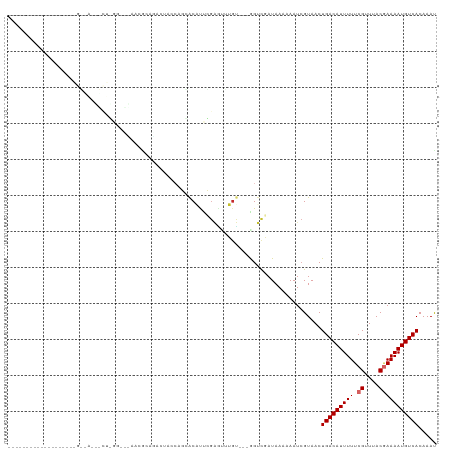
>dm3.chr3R 13897355 97 - 27905053 ------------------GGGAAACUUUGGACAGAACGGAUACCAGC-GCACAUCCGAGUUUGU---GGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAC ------------------((....))..........(((((......-....))))).((((((---(((((((.....))))..)))((((((((.((....)))))))))))))))) ( -22.20, z-score = -0.94, R) >droSim1.chr3R 19923172 98 + 27517382 ------------------GGGAAACUCUGGGCAGAACGGAUACUACCAGCACAUCCGAGUUUGU---GGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAC ------------------.(((....((((..((........)).))))....)))..((((((---(((((((.....))))..)))((((((((.((....)))))))))))))))) ( -24.20, z-score = -1.65, R) >droSec1.super_12 1854574 98 + 2123299 ------------------GGGAAACUCUGGGCAGAACGGAUACUACCAGCACAUCCGAGUUUGU---GGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAC ------------------.(((....((((..((........)).))))....)))..((((((---(((((((.....))))..)))((((((((.((....)))))))))))))))) ( -24.20, z-score = -1.65, R) >droYak2.chr3R 2490923 83 - 28832112 ---------------------------------UUGGAAAAAAUACCAGCACAUUCGAAUUUGU---AGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAU ---------------------------------((((........)))).........((((((---(((((((.....))))..)))((((((((.((....)))))))))))))))) ( -16.10, z-score = -1.89, R) >droEre2.scaffold_4770 10008229 80 + 17746568 ------------------------------------GGAAGAAGACCAGCACAUCCGAGUUUGU---AGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAU ------------------------------------(((.(..........).)))..((((((---(((((((.....))))..)))((((((((.((....)))))))))))))))) ( -15.80, z-score = -1.35, R) >droWil1.scaffold_181089 7563304 80 + 12369635 ----------------------------------GAUGCGCGCUGGCCCUGCAUUU--UCUUCG---GGCCAAUAAAAAAUAGUAACUGACAUUUUUCGUUUACAAAAAUGUCACAAAU ----------------------------------.........((((((.......--.....)---)))))...............((((((((((.(....)))))))))))..... ( -18.70, z-score = -2.05, R) >droVir3.scaffold_13047 12441107 96 + 19223366 AACUUGAACUUCAUC---AGUGCCUCCACGG-----------------CGGCACUGGGGAGUGC---AGCUAAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAU ...(((.(((((.((---((((((.(....)-----------------.))))))))))))).)---))..................(((((((((.((....)))))))))))..... ( -31.10, z-score = -4.33, R) >droMoj3.scaffold_6540 31513171 116 + 34148556 AACGUGAACUUCAUC---AGCGACGGCUCGACUCGACGCGGCUCGGCUCGACGCUGGGCAGCGCUGGGGCUAAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAU ...((((........---(((..(((((((...))).((.(((((((.....))))))).))))))..))).........((((((.(((......))).)))))).....)))).... ( -35.90, z-score = -2.00, R) >droGri2.scaffold_14830 3610386 114 + 6267026 AACUUCAACUUCAGCUCGAACUGCAGCUUGGACUUCAUCAGUUUG--UCGGCUCUGGGGAGUGC---AGCUAAUAAAAAAUCGUAACUGACAUUUUUCGUUUACCAAAAUGUCACAAAU .............((.......))((((.(.(((((.((((...(--....).))))))))).)---))))................(((((((((.........)))))))))..... ( -21.60, z-score = 0.10, R) >consensus ___________________G__A___CU_GG___AACGCAGACUACCAGCACAUUCGAGUUUGU___GGUCGAUAAAAAAUCGUAACUGACAUUUUUCGUUUACGAAAAUGUCACAAAU .......................................................................................(((((((((.((....)))))))))))..... ( -8.71 = -8.93 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:23 2011