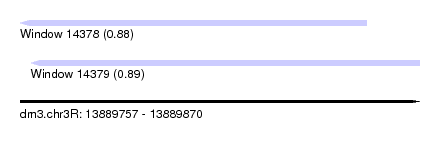
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,889,757 – 13,889,870 |
| Length | 113 |
| Max. P | 0.889861 |

| Location | 13,889,757 – 13,889,855 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.40086 |
| G+C content | 0.40489 |
| Mean single sequence MFE | -14.10 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
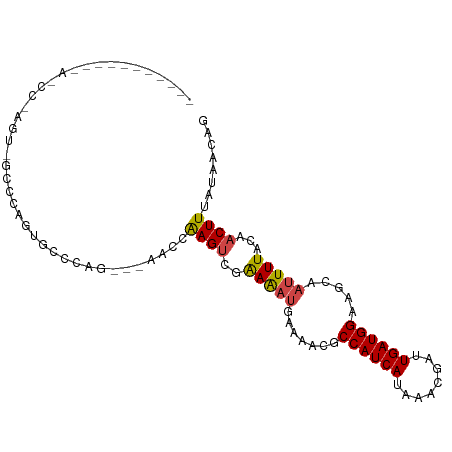
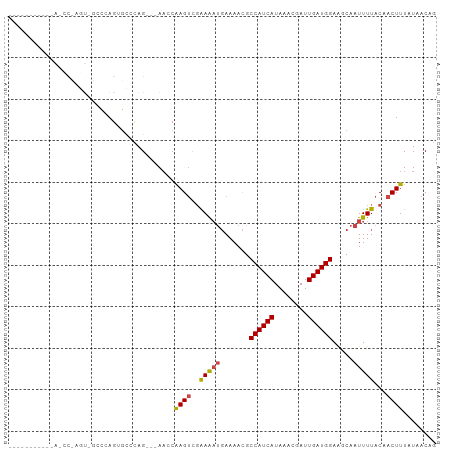
>dm3.chr3R 13889757 98 - 27905053 AGCCCCCAAGAA-CCCAGU-GCCCAGUGCCCAG---AACCAAGUUGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAACAG ............-....(.-.(...)..)....---....(((((((((((.......((((((........)))))).....))))).))))))........ ( -15.00, z-score = -1.92, R) >droYak2.chr3R 2483545 79 - 28832112 ---------------------ACGAGAGCCUAG---AACCGAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAACCAAUUUUACAACUUUAUAACAC ---------------------............---....((((.((((((.......((((((........)))))).....))))).).))))........ ( -11.10, z-score = -1.82, R) >droEre2.scaffold_4770 10000854 79 + 17746568 ---------------------CCAAGAGCCCAG---AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAACAG ---------------------............---....((((.((((((.......((((((........)))))).....))))).).))))........ ( -10.70, z-score = -1.92, R) >droSec1.super_12 1847211 100 + 2123299 AGCCUCCAAGAACCCCAGUGGCCCAGUGCCCAG---ACCCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAACAG .((.((((.........(.(((.....))))..---.....(((((...((((........))))...)))))..)))).))..................... ( -16.20, z-score = -1.20, R) >droSim1.chr3R 19915841 90 + 27517382 --------AGAA-CCUAGU-GCCCAGUGCCCAG---AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAACAG --------....-.((.(.-((.....))).))---....((((.((((((.......((((((........)))))).....))))).).))))........ ( -11.20, z-score = -0.42, R) >droAna3.scaffold_13340 11865664 92 + 23697760 -----------AGUCGAAUGCGAAAGCUUUCAGUAAAACCAAGCCCGAGUCCAAAAUGCCAUCAUAAACGAUUGAUGGAGGCAAUUUUACAACUUUAUAACAG -----------....(((.((....)).))).((((((....(((.(....)......((((((........)))))).)))..))))))............. ( -20.40, z-score = -2.62, R) >consensus ___________A_CC_AGU_GCCCAGUGCCCAG___AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAACAG ........................................((((..(((((.......((((((........)))))).....)))))...))))........ ( -9.47 = -9.55 + 0.08)

| Location | 13,889,760 – 13,889,870 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.46789 |
| G+C content | 0.42443 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
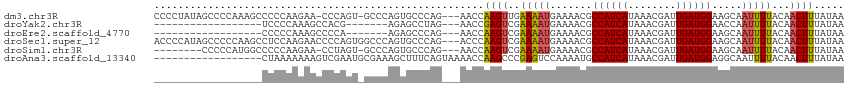
>dm3.chr3R 13889760 110 - 27905053 CCCCUAUAGCCCCAAAGCCCCCAAGAA-CCCAGU-GCCCAGUGCCCAG---AACCAAGUUGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAA ...........................-....(.-.(...)..)....---....(((((((((((.......((((((........)))))).....))))).))))))..... ( -15.00, z-score = -1.66, R) >droYak2.chr3R 2483548 87 - 28832112 ------------------UCCCCAAAGCCACG-------AGAGCCUAG---AACCGAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAACCAAUUUUACAACUUUAUAA ------------------..............-------.........---....((((.((((((.......((((((........)))))).....))))).).))))..... ( -11.10, z-score = -1.18, R) >droEre2.scaffold_4770 10000857 87 + 17746568 ------------------CCCCCAAAGCCCCA-------AGAGCCCAG---AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAA ------------------..............-------.........---....((((.((((((.......((((((........)))))).....))))).).))))..... ( -10.70, z-score = -1.58, R) >droSec1.super_12 1847214 112 + 2123299 ACCCCAUAGCCCCCAAGCCUCCAAGAACCCCAGUGGCCCAGUGCCCAG---ACCCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAA ................((.((((.........(.(((.....))))..---.....(((((...((((........))))...)))))..)))).)).................. ( -16.20, z-score = -1.09, R) >droSim1.chr3R 19915844 102 + 27517382 --------CCCCCAUGGCCCCCAAGAA-CCUAGU-GCCCAGUGCCCAG---AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAA --------....((((((.(.......-....).-)))..))).....---....((((.((((((.......((((((........)))))).....))))).).))))..... ( -12.20, z-score = 0.36, R) >droAna3.scaffold_13340 11865667 97 + 23697760 ------------------CUAAAAAAAGUCGAAUGCGAAAGCUUUCAGUAAAACCAAGCCCGAGUCCAAAAUGCCAUCAUAAACGAUUGAUGGAGGCAAUUUUACAACUUUAUAA ------------------......(((((.(((.((....)).))).((((((....(((.(....)......((((((........)))))).)))..)))))).))))).... ( -22.30, z-score = -3.29, R) >consensus __________________CCCCAAAAACCCCAGU_GCCCAGUGCCCAG___AACCAAGUCGAAAAUGAAAACGCCAUCAUAAACGAUUGAUGGAAGCAAUUUUACAACUUUAUAA .......................................................((((..(((((.......((((((........)))))).....)))))...))))..... ( -9.47 = -9.55 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:17 2011