
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,857,796 – 13,857,900 |
| Length | 104 |
| Max. P | 0.932144 |

| Location | 13,857,796 – 13,857,900 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Shannon entropy | 0.25691 |
| G+C content | 0.41189 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
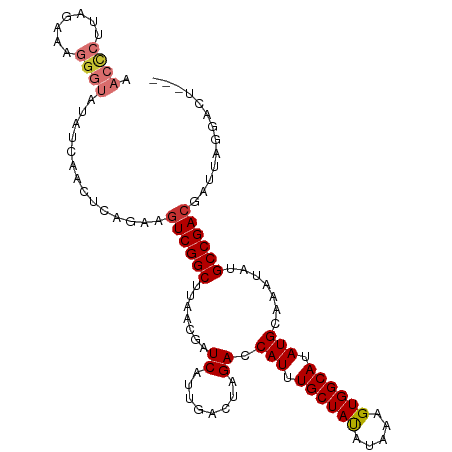
>dm3.chr3R 13857796 104 + 27905053 AACCCUCGGAAAGGGUAUAUCAACUCAGAAAUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU--- .(((((.....)))))........((((..((((......))))..)))).....((....(((......)))(((((((....)))))))........))...--- ( -23.90, z-score = -1.53, R) >droSim1.chr3R 19889612 104 - 27517382 AACCCUUAGAAAGGGUAUAUCAACUCAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU--- .(((((.....)))))........((((..((((......))))..)))).....((....(((......)))(((((((....)))))))........))...--- ( -24.30, z-score = -1.66, R) >droSec1.super_12 1823916 104 - 2123299 AACCCUAGGAAAGGGUAUAUCAACUCAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU--- ...(((((.....(((((((....((((..((((......))))..))))......(((.((((((.....)))))).)))...))))))).....)))))...--- ( -25.60, z-score = -1.86, R) >droYak2.chr3R 2453560 104 + 28832112 UACCCAACGAUAGGGUAUAUAACCUCAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUACAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU--- (((((.......))))).....(((.(...((((((.......((........)).(((.((((((.....)))))).))).......))))))..).)))...--- ( -28.30, z-score = -3.12, R) >droEre2.scaffold_4770 9977675 104 - 17746568 AACCCUACGAAAGGGUAUAUCAACUCAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUACAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU--- ...(((((....)(((((((....((((..((((......))))..))))......(((.((((((.....)))))).)))...)))))))......))))...--- ( -27.60, z-score = -2.90, R) >droAna3.scaffold_13340 11842922 107 - 23697760 AACCCGUUUAAAGAGUAUCAGAAGUCAAAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGCCUAGU .....((((((..((((.....((((((..((((......))))..))))))........)))).......(((((((((....))))))).))..))))))..... ( -25.22, z-score = -1.81, R) >dp4.chr2 28913403 103 + 30794189 UGCUCUUACACCUGAGAC-UCACCGUAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUGGCAAC--- ((((((((....))))).-...........((((((.......((........)).(((.((((((.....)))))).))).......)))))).....)))..--- ( -22.00, z-score = -0.74, R) >droPer1.super_6 4240102 103 + 6141320 UGCUCUUACACCUGAGAC-UCACCGUAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUGGCAAC--- ((((((((....))))).-...........((((((.......((........)).(((.((((((.....)))))).))).......)))))).....)))..--- ( -22.00, z-score = -0.74, R) >consensus AACCCUUAGAAAGGGUAUAUCAACUCAGAAGUCGGCUUAACGAUCAUUGACUAGACCAUUUGCUAUAUAAAGUGGCAUAUGCAAAUAUGCCGACGAUUAGGACU___ .((((.......))))..............((((((.......((........)).(((.((((((.....)))))).))).......))))))............. (-18.74 = -19.40 + 0.66)


| Location | 13,857,796 – 13,857,900 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Shannon entropy | 0.25691 |
| G+C content | 0.41189 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13857796 104 - 27905053 ---AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGAUUUCUGAGUUGAUAUACCCUUUCCGAGGGUU ---.(((((....((((((..(((((((((((......))))).))))))((((........)))).....)))))).....))..)))..((((((...)))))). ( -26.20, z-score = -1.69, R) >droSim1.chr3R 19889612 104 + 27517382 ---AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUGAGUUGAUAUACCCUUUCUAAGGGUU ---..........((((((..(((((((((((......))))).))))))((((........)))).....))))))..............(((((.....))))). ( -27.70, z-score = -2.40, R) >droSec1.super_12 1823916 104 + 2123299 ---AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUGAGUUGAUAUACCCUUUCCUAGGGUU ---..........((((((..(((((((((((......))))).))))))((((........)))).....))))))..............(((((.....))))). ( -28.20, z-score = -2.52, R) >droYak2.chr3R 2453560 104 - 28832112 ---AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUGUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUGAGGUUAUAUACCCUAUCGUUGGGUA ---...(((....((((((....(((((((........)))))))..(((((((........)))))))..)))))).....))).....(((((.......))))) ( -30.80, z-score = -2.71, R) >droEre2.scaffold_4770 9977675 104 + 17746568 ---AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUGUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUGAGUUGAUAUACCCUUUCGUAGGGUU ---..........((((((....(((((((........)))))))..(((((((........)))))))..))))))..............(((((.....))))). ( -28.60, z-score = -2.30, R) >droAna3.scaffold_13340 11842922 107 + 23697760 ACUAGGCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUUUGACUUCUGAUACUCUUUAAACGGGUU ....(((((....((((((..(((((((((((......))))).))))))((((........)))).....))))))..........(((......)))...))))) ( -24.80, z-score = -1.43, R) >dp4.chr2 28913403 103 - 30794189 ---GUUGCCAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUACGGUGA-GUCUCAGGUGUAAGAGCA ---.(..((....((((((..(((((((((((......))))).))))))((((........)))).....))))))......))..)-(.(((........)))). ( -26.90, z-score = -1.02, R) >droPer1.super_6 4240102 103 - 6141320 ---GUUGCCAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUACGGUGA-GUCUCAGGUGUAAGAGCA ---.(..((....((((((..(((((((((((......))))).))))))((((........)))).....))))))......))..)-(.(((........)))). ( -26.90, z-score = -1.02, R) >consensus ___AGUCCUAAUCGUCGGCAUAUUUGCAUAUGCCACUUUAUAUAGCAAAUGGUCUAGUCAAUGAUCGUUAAGCCGACUUCUGAGUUGAUAUACCCUUUCUAAGGGUU .............((((((..(((((((((((......))))).))))))((((........)))).....))))))..............((((.......)))). (-23.21 = -23.40 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:15 2011