
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,856,472 – 13,856,611 |
| Length | 139 |
| Max. P | 0.682534 |

| Location | 13,856,472 – 13,856,611 |
|---|---|
| Length | 139 |
| Sequences | 7 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.55089 |
| G+C content | 0.42855 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -14.06 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
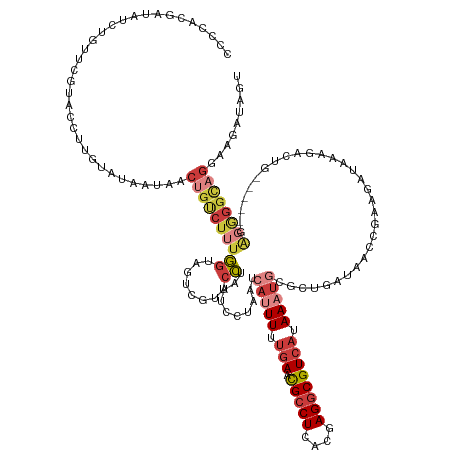
>dm3.chr3R 13856472 139 + 27905053 CCCCACGAUAUCAGUUCGUACCUUGUAUAAUAACUGUCUUUGGUAGUCGUUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUGAAAACCGUAGAUAAAGACUG-----AGGAGGCAGGAAGAUAGU ....((((.......))))..............((((((((..(((((....(((........(((((.(((.(((((....)))))))).)))))((.(....).))))).....)))))-----.))))))))......... ( -33.00, z-score = -0.63, R) >droWil1.scaffold_181089 7537029 120 - 12369635 CCCCACCGCACCACUUGUCACUUUCUACA-CAUUUUUCCGUUUGAAUAUUGCAAAAUUCAAAUCAAUUUCAAAUGGCUGACGAGGCGUCAUAAACACGCCUUUAAAACAAAAUAGACAAGA----------------------- .............((((((..........-.......(((((((((.((((............))))))))))))).....(((((((.......)))))))............)))))).----------------------- ( -26.20, z-score = -3.91, R) >droEre2.scaffold_4770 9976371 137 - 17746568 CCCCACGAAAUGUAUUCGUACCUUGUGUAAUAACUGUCUUUGGAAGUCGCUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUCAUG-CCGAAGAUAAAGACU------GAGGGGCAGGAAGAUAGU ....(((((.....))))).............(((((((((.(....)((((((..((.....(((((.(((.(((((....)))))))).))))).((....)-)..........))..------.))))))..))))))))) ( -35.70, z-score = -0.95, R) >droYak2.chr3R 2452217 143 + 28832112 CCCGACUAAAUCCAUUUGUACCUUGUAUAAUAACUGCCUUUGGAAGUCGUUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUCAUA-CCGGAGAUAAAGACUGAGACUGAGGGGCAGGAAGAUAGU ....((((.......((((((...))))))...((((((((((.((((.((((..........(((((.(((.(((((....)))))))).)))))........-..)))).....))))....)).)))))))).....)))) ( -35.65, z-score = -0.77, R) >droSec1.super_12 1822644 141 - 2123299 CCCCACGAUAUCUGUUCGUACCUUGUAUAAUAACUGUCUUUGGUAGUCGUUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUGAAAACCGUAGAUAAAUACUGUG---GAGGGGCAGGAAGAUAGU ....((((.......)))).............(((((((((....(((.((((..........(((((.(((.(((((....)))))))).)))))........((.(((......))).)))---))).)))..))))))))) ( -35.30, z-score = -0.68, R) >droSim1.chr3R 19888348 141 - 27517382 CCCCGCGAUAUCUGUUCGUACCUUGUAUAAUAACUGUCUUUGGUAGUCGUUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUGAAAACCGUAGAUAAAGACUGAG---GAGGGGCAGGAAGAUAGU ....((((.......)))).............(((((((((....(((.(((((..((...(((.(((.(((.(((((....)))))))).)))((((.(....).)))))))...))...))---))).)))..))))))))) ( -36.90, z-score = -0.72, R) >droAna3.scaffold_13340 11841873 114 - 23697760 ----------------------UUGGAUAAUAACUGUCUUUGGCAGUCGUUCCUAAUCCUAAUCAAUUUUGAACGCCUCACGAGGCGUCAUAAAUACGUUUUUU-CGAAAGAUAAAGACGG-----GAUGGGUCG-AGGAUAG- ----------------------...........(((((((((((...((((((............(((((((.(((((....)))))))).))))..(((((((-(....)).))))))))-----)))).))))-)))))))- ( -38.20, z-score = -3.84, R) >consensus CCCCACGAUAUCUGUUCGUACCUUGUAUAAUAACUGUCUUUGGUAGUCGUUCCUAAUCCUAAUCAUUUUUGAACGCCUCACGAGGCGUCAUAAAUGCGCUGAUAACCGAAGAUAAAGACUG_____GAGGGGCAGGAAGAUAGU .................................((((((((((........))..........(((((.(((.(((((....)))))))).)))))...............................))))))))......... (-14.06 = -16.00 + 1.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:12 2011