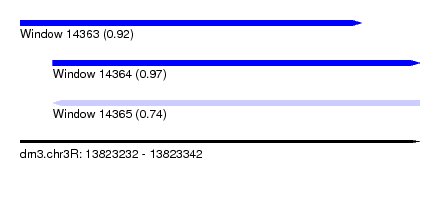
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,823,232 – 13,823,342 |
| Length | 110 |
| Max. P | 0.973576 |

| Location | 13,823,232 – 13,823,326 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Shannon entropy | 0.47037 |
| G+C content | 0.40442 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
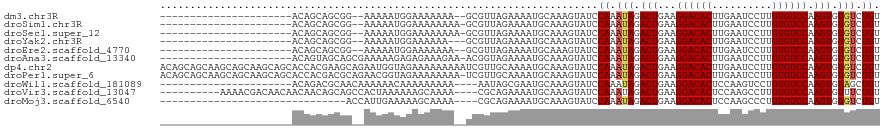
>dm3.chr3R 13823232 94 + 27905053 ----------------------ACAGCAGCGG--AAAAAUGGAAAAAAA--GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((--(....((((......--(((((....)))))......))))..((.(((...((((((..(....)..)))))).))).))))))) ( -23.40, z-score = -2.46, R) >droSim1.chr3R 19851883 95 - 27517382 ----------------------ACAGCAGCGG--AAAAAUGGAAAAAAAA-GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((--(....((((.......-(((((....)))))......))))..((.(((...((((((..(....)..)))))).))).))))))) ( -22.82, z-score = -2.26, R) >droSec1.super_12 1790151 95 - 2123299 ----------------------ACAGCAGCGG--AAAAAUGGAAAAAAAA-GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((--(....((((.......-(((((....)))))......))))..((.(((...((((((..(....)..)))))).))).))))))) ( -22.82, z-score = -2.26, R) >droYak2.chr3R 2415905 93 + 28832112 ----------------------ACAGCAGCGG--AAAAAUGGAAAAAA---GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((--(....((((.....---(((((....)))))......))))..((.(((...((((((..(....)..)))))).))).))))))) ( -23.00, z-score = -2.29, R) >droEre2.scaffold_4770 9941465 94 - 17746568 ----------------------ACAGCAGCGG--AAAAAUGGAAAAAAA--GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((--(....((((......--(((((....)))))......))))..((.(((...((((((..(....)..)))))).))).))))))) ( -23.40, z-score = -2.46, R) >droAna3.scaffold_13340 11805308 97 - 23697760 ----------------------ACAGUAGCAGCGAAAAAGAGAGAAAGAA-ACGGUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ----------------------......((((...............((.-((.(((.....)))...)).))...(((.(((...((((((..(....)..)))))).))).))))))) ( -18.00, z-score = -0.67, R) >dp4.chr2 28865968 120 + 30794189 ACAGCAGCAAGCAGCAAGCAGCACCACGAAGCAGAAUGGUAGAAAAAAAAAUCGUUGCAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ...(((((.....))..(((((((((..........)))).((........)))))))....)))........((.(((.(((...((((((..(....)..)))))).))).))).)). ( -27.80, z-score = -0.71, R) >droPer1.super_6 4192714 119 + 6141320 ACAGCAGCAAGCAGCAAGCAGCACCACGACGCAGAACGGUAGAAAAAAAA-UCGUUGCAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU ...((.((.........)).)).....((((((...((((..........-...((((.....)))).............))))..((((((..(....)..))))))...))))))... ( -28.27, z-score = -0.60, R) >droWil1.scaffold_181089 7500194 94 - 12369635 ----------------------ACAGACGCAACAAAAAACAAAAAAAAA----AAUAGCGAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGUCCUGUGUCCAAGUGUAGCUGU ----------------------...........................----.((((((.((((...)))).)...((.(((...((((((..........)))))).))).))))))) ( -16.80, z-score = -1.69, R) >droVir3.scaffold_13047 6861113 106 + 19223366 ----------AAAACGACAACAACAACAGCAGCCACUAAAAAAGCAAAA----CGCAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGCCUUGUGUCCAAGUGUUUCUGU ----------...............((((.(((.(((............----.(((.....))).....................((((((..........)))))).)))))).)))) ( -19.30, z-score = -1.01, R) >droMoj3.scaffold_6540 2725129 85 + 34148556 -------------------------------ACCAUUGAAAAAGCAAAA----CGCAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGCCCUGUGUCCAAGUGUGUCUGU -------------------------------..................----.(((.....)))........((.(((.(((...((((((..........)))))).))).))).)). ( -16.80, z-score = -1.40, R) >consensus ______________________ACAGCAGCGG__AAAAAUAGAAAAAAA__GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGU .........................................................................((.(((.(((...((((((..........)))))).))).))).)). (-13.37 = -13.48 + 0.11)
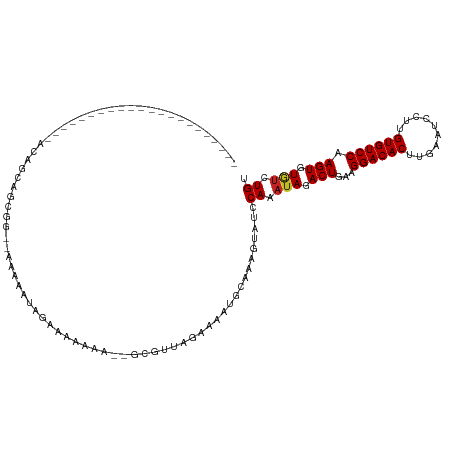
| Location | 13,823,241 – 13,823,342 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.51167 |
| G+C content | 0.42153 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13823241 101 + 27905053 ----GAAAAAUGGAAAAAAA--GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCAGCUUC----------- ----(((...((((......--(((((....)))))......)))).((((((....((((((..(....)..))))))......)))))).......)))......----------- ( -22.50, z-score = -2.12, R) >droSim1.chr3R 19851892 102 - 27517382 ----GAAAAAUGGAAAAAAAA-GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCAGCUUC----------- ----(((...((((.......-(((((....)))))......)))).((((((....((((((..(....)..))))))......)))))).......)))......----------- ( -21.92, z-score = -1.99, R) >droSec1.super_12 1790160 102 - 2123299 ----GAAAAAUGGAAAAAAAA-GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCAGCUUC----------- ----(((...((((.......-(((((....)))))......)))).((((((....((((((..(....)..))))))......)))))).......)))......----------- ( -21.92, z-score = -1.99, R) >droYak2.chr3R 2415914 100 + 28832112 ----GAAAAAUGGAAAAAA---GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCGGCUUC----------- ----(((...((((.....---(((((....)))))......)))).((((((....((((((..(....)..))))))......)))))).......)))......----------- ( -22.10, z-score = -1.70, R) >droEre2.scaffold_4770 9941474 101 - 17746568 ----GAAAAAUGGAAAAAAA--GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCAGCUUC----------- ----(((...((((......--(((((....)))))......)))).((((((....((((((..(....)..))))))......)))))).......)))......----------- ( -22.50, z-score = -2.12, R) >droAna3.scaffold_13340 11805318 103 - 23697760 ---CGAAAAAGAGAGAAAGAA-ACGGUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCUUCUCUGGAGCUUC----------- ---........((((((.((.-((.(((.....)))...))...((.(((.(((...((((((..(....)..)))))).))).))).)))).))))))........----------- ( -23.50, z-score = -1.01, R) >dp4.chr2 28865997 118 + 30794189 AGCAGAAUGGUAGAAAAAAAAAUCGUUGCAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCGGUCCCGCUUGUCCCUCUUCUGCUCG (((((((.((................(((.....)))((((..((..((((((....((((((..(....)..))))))......)))))).))....))))...))..))))))).. ( -29.90, z-score = -2.57, R) >droPer1.super_6 4192743 117 + 6141320 CGCAGAACGGUAGAAAAAAAA-UCGUUGCAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCGGUCCCGCUUGUCCCUCUUCUGCUCG .((((((.((...........-....(((.....)))((((..((..((((((....((((((..(....)..))))))......)))))).))....))))...))..))))))... ( -29.50, z-score = -2.20, R) >droWil1.scaffold_181089 7500205 108 - 12369635 ----AAAAAACAAAAAAAAAA----AUAGCGAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGUCCUGUGUCCAAGUGUAGCUGUCCUU-UCGUCAUUCCGCCCCCGCCUA- ----.................----...((((((((((((....((..((.(((...((((((..........)))))).))).))..))..)))-).).)))).))).........- ( -20.40, z-score = -3.04, R) >droVir3.scaffold_13047 6861129 108 + 19223366 CAGCAGCCACUAAAAAAGCAAAACG-CAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGCCUUGUGUCCAAGUGUUUCUGUUCUGCCCAGCACAGCGCA--------- ..((...((((.............(-((.....))).....................((((((..........)))))).))))...((((.((....)).)))).)).--------- ( -22.00, z-score = -0.01, R) >droMoj3.scaffold_6540 2725130 86 + 34148556 ------CCAUUGAAAAAGCAAAACG-CAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUCCAAGCCCUGUGUCCAAGUGUGUCUGUCC------------------------- ------..................(-((.....)))...........((((((....((((((..........))))))......))))))..------------------------- ( -16.90, z-score = -1.64, R) >consensus ____GAAAAAUGGAAAAAAAA_GCGUUAGAAAAUGCAAAGUAUCCAAAUAGACUGAAGGACACUUGAAUCCUUGUGUCCAAGUGUGUCUGUCCCUCUCUUCAGCUUC___________ ............................................((.(((.(((...((((((..........)))))).))).))).))............................ (-13.37 = -13.48 + 0.11)
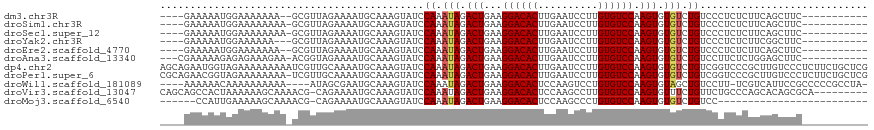

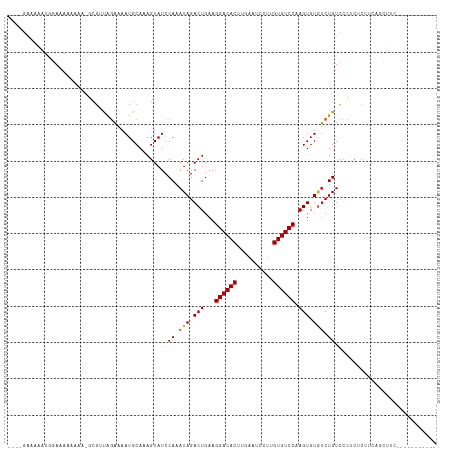
| Location | 13,823,241 – 13,823,342 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.51167 |
| G+C content | 0.42153 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -14.24 |
| Energy contribution | -13.51 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13823241 101 - 27905053 -----------GAAGCUGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC--UUUUUUUCCAUUUUUC---- -----------(((((.(((((((((((((......((((....))))........)))))))).((((....))))........)))))....))--))).............---- ( -22.64, z-score = -0.92, R) >droSim1.chr3R 19851892 102 + 27517382 -----------GAAGCUGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC-UUUUUUUUCCAUUUUUC---- -----------(((((.(((((((((((((......((((....))))........)))))))).((((....))))........)))))....))-)))..............---- ( -22.64, z-score = -0.90, R) >droSec1.super_12 1790160 102 + 2123299 -----------GAAGCUGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC-UUUUUUUUCCAUUUUUC---- -----------(((((.(((((((((((((......((((....))))........)))))))).((((....))))........)))))....))-)))..............---- ( -22.64, z-score = -0.90, R) >droYak2.chr3R 2415914 100 - 28832112 -----------GAAGCCGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC---UUUUUUCCAUUUUUC---- -----------......((((((((.(..(((....((((....)))).....((((((((............)))))))).......)))..).)---)))))))........---- ( -22.20, z-score = -0.93, R) >droEre2.scaffold_4770 9941474 101 + 17746568 -----------GAAGCUGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC--UUUUUUUCCAUUUUUC---- -----------(((((.(((((((((((((......((((....))))........)))))))).((((....))))........)))))....))--))).............---- ( -22.64, z-score = -0.92, R) >droAna3.scaffold_13340 11805318 103 + 23697760 -----------GAAGCUCCAGAGAAGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUACCGU-UUCUUUCUCUCUUUUUCG--- -----------((((....(((((((((..(((...((((....)))).....((((((((............)))))))).............))-))))))))))...)))).--- ( -25.80, z-score = -1.75, R) >dp4.chr2 28865997 118 - 30794189 CGAGCAGAAGAGGGACAAGCGGGACCGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUGCAACGAUUUUUUUUUCUACCAUUCUGCU ..((((((((((((((..(((....)).)......(((((((........))))))))))))))........(((.....((((.....)))).((........))..)))))))))) ( -32.10, z-score = -2.33, R) >droPer1.super_6 4192743 117 - 6141320 CGAGCAGAAGAGGGACAAGCGGGACCGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUGCAACGA-UUUUUUUUCUACCGUUCUGCG ...(((((((((((((..(((....)).)......(((((((........)))))))))))))).((((....)))).(.((((.....)))).).-..............)))))). ( -31.00, z-score = -1.71, R) >droWil1.scaffold_181089 7500205 108 + 12369635 -UAGGCGGGGGCGGAAUGACGA-AAGGACAGCUACACUUGGACACAGGACUUGGAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUCGCUAU----UUUUUUUUUUGUUUUUU---- -...((((((.(.(((((((..-(((((((.(((..((((....))))...)))..)))))))..))).)))).)...)))))).........----.................---- ( -28.80, z-score = -2.16, R) >droVir3.scaffold_13047 6861129 108 - 19223366 ---------UGCGCUGUGCUGGGCAGAACAGAAACACUUGGACACAAGGCUUGGAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUG-CGUUUUGCUUUUUUAGUGGCUGCUG ---------.((((..(...((((((((((((((..((((....))))((...((((((((............)))))))).))..))))).-.))))))))).....)..)).)).. ( -33.60, z-score = -1.17, R) >droMoj3.scaffold_6540 2725130 86 - 34148556 -------------------------GGACAGACACACUUGGACACAGGGCUUGGAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUG-CGUUUUGCUUUUUCAAUGG------ -------------------------.(.((((....((((....))))((...((((((((............)))))))).))....))))-)..................------ ( -18.30, z-score = -0.14, R) >consensus ___________GAAGCUGAAGAGAGGGACAGACACACUUGGACACAAGGAUUCAAGUGUCCUUCAGUCUAUUUGGAUACUUUGCAUUUUCUAACGC_UUUUUUUUCCAUUUUUC____ .................(((((...(((........((((....)))).....((((((((............)))))))).......................))).)))))..... (-14.24 = -13.51 + -0.73)

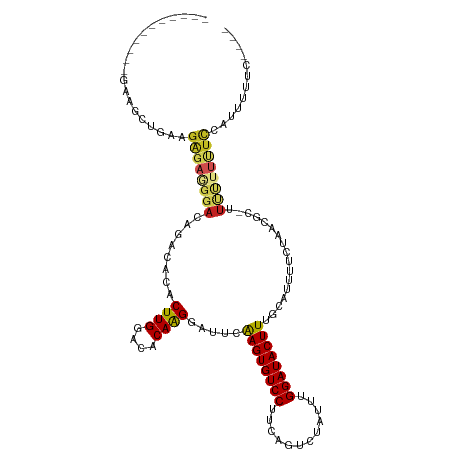
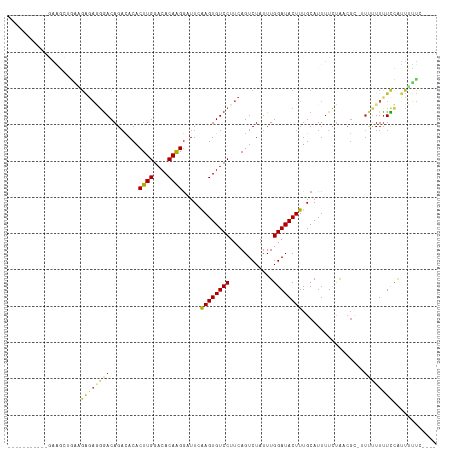
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:24:05 2011