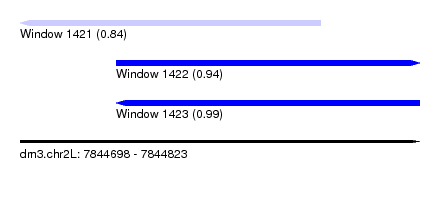
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,844,698 – 7,844,823 |
| Length | 125 |
| Max. P | 0.988799 |

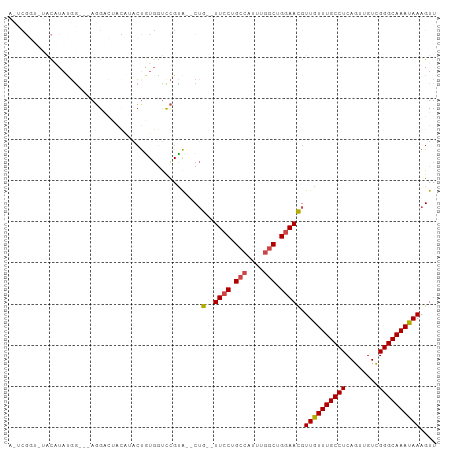
| Location | 7,844,698 – 7,844,792 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Shannon entropy | 0.17024 |
| G+C content | 0.44437 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7844698 94 - 23011544 ------GUCCGUA--CUG--UUCCUGCCAUUUGGCUGGAAUGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA ------...((((--(.(--((((.(((....))).))))).((((((((((........)))))))))).......(((((.......)))))..)))))... ( -29.60, z-score = -2.63, R) >droSim1.chr2L 7641970 94 - 22036055 ------GUCCGUA--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA ------...((((--(.(--((((.(((....))).))))).((((((((((........)))))))))).......(((((.......)))))..)))))... ( -31.70, z-score = -3.11, R) >droSec1.super_3 3359026 94 - 7220098 ------GUCCGCA--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA ------...((((--(.(--((((.(((....))).))))).((((((((((........)))))))))).......(((((.......)))))..)))))... ( -33.70, z-score = -3.41, R) >droYak2.chr2L 17272157 94 + 22324452 ------GUCCGUG--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA ------...((..--(.(--((((.(((....))).))))).((((((((((........)))))))))).......(((((.......)))))..)..))... ( -30.90, z-score = -2.59, R) >droEre2.scaffold_4929 16766316 100 - 26641161 GUCUGUACUCGCA--CAG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA ........(((((--(((--((((.(((....))).))))).((((((((((........)))))))))).......(((((.......))))).))))))).. ( -36.60, z-score = -3.68, R) >droAna3.scaffold_12943 3579432 98 + 5039921 ------GUUUGUAGUCUGGCUUGCUGGCUUCUCGCUGGAAUGUUAUUUGCCUCAGUUGUUGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCCAUA ------..........((((...(..((.....))..)(((.((((((((((........)))))))))).)))...(((((.......)))))....)))).. ( -23.50, z-score = 0.05, R) >consensus ______GUCCGUA__CUG__UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAUAGAUAGAGACAUGUGCGAUA .........((((.......((((.(((....))).))))..((((((((((........)))))))))).......(((((.......)))))...))))... (-21.43 = -21.60 + 0.17)

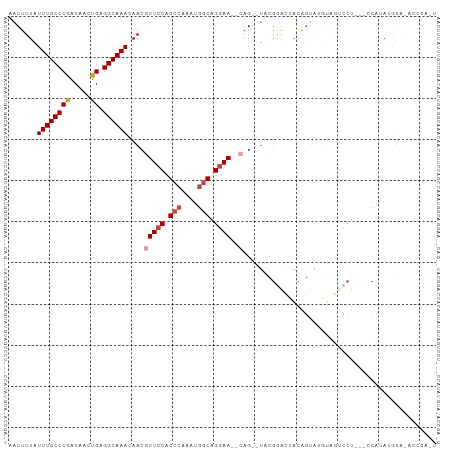
| Location | 7,844,728 – 7,844,823 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Shannon entropy | 0.43872 |
| G+C content | 0.47244 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -12.37 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
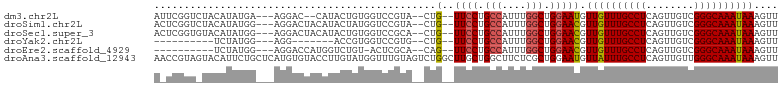
>dm3.chr2L 7844728 95 + 23011544 AACUUUAUUUGCCCGACAACUGAGGCAAACAACAUUCCAGCCAAAUGGCAGGAA--CAG--UACGGACCACAGUAUG--GUCCU---UCAUAUGUAGACCGAAU .......((((((((.....)).))))))..(((((((.(((....))).))))--...--...((((((.....))--)))).---.....)))......... ( -25.30, z-score = -2.37, R) >droSim1.chr2L 7642000 97 + 22036055 AACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAA--CAG--UACGGACCAUAGUAUGUAGUCCU---CCAUAUGUAGACCGAGU .(((...((((((((.....)).))))))....(((((.(((....))).))))--)))--).(((......(((((.......---.))))).....)))... ( -25.30, z-score = -1.85, R) >droSec1.super_3 3359056 97 + 7220098 AACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAA--CAG--UGCGGACCACAGUAUGUAGUCCU---CCAUAUGUACACCGAGU .((((..((((((((.....)).))))))....(((((.(((....))).))))--).(--(((((((.((.....)).)))).---......))))...)))) ( -27.81, z-score = -2.13, R) >droYak2.chr2L 17272187 80 - 22324452 AACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAA--CAG--CACGGACCACGGU-------CCU---CCAUAGA---------- .......((((((((.....)).))))))....(((((.(((....))).))))--)..--...((((....))-------)).---.......---------- ( -26.00, z-score = -3.48, R) >droEre2.scaffold_4929 16766346 86 + 26641161 AACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAA--CUG--UGCGAGU-ACAGACCAUGGUCCU---CCAUAGA---------- .......((((((((.....)).))))))....(((((.(((....))).))))--)((--((.(((.-...(((....)))))---)))))..---------- ( -26.00, z-score = -2.44, R) >droAna3.scaffold_12943 3579462 104 - 5039921 AACUUUAUUUGCCCAACAACUGAGGCAAAUAACAUUCCAGCGAGAAGCCAGCAAGCCAGACUACAAACCAUACAAGGUACACAUGAGCAGAAUGUACUACGGUU .....((((((((((.....)).))))))))((((((..((.........))..((((........(((......))).....)).)).))))))((....)). ( -21.32, z-score = -1.84, R) >consensus AACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAA__CAG__UACGGACCACAGUAUGUAGUCCU___CCAUAUGUA_ACCGA_U .......((((((((.....)).)))))).....((((.(((....))).)))).................................................. (-12.37 = -12.73 + 0.36)

| Location | 7,844,728 – 7,844,823 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Shannon entropy | 0.43872 |
| G+C content | 0.47244 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
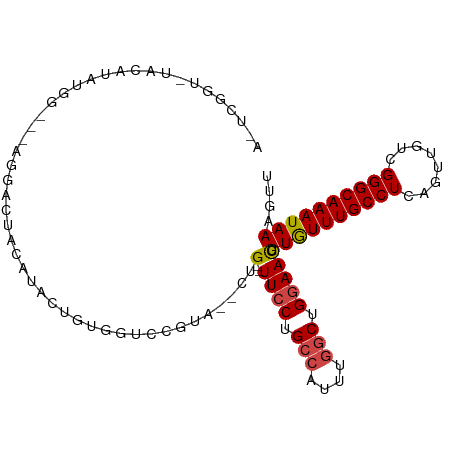
>dm3.chr2L 7844728 95 - 23011544 AUUCGGUCUACAUAUGA---AGGAC--CAUACUGUGGUCCGUA--CUG--UUCCUGCCAUUUGGCUGGAAUGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ...(((.(((((((((.---.....--)))).))))).)))..--..(--((((.(((....))).))))).((((((((((........)))))))))).... ( -34.30, z-score = -3.56, R) >droSim1.chr2L 7642000 97 - 22036055 ACUCGGUCUACAUAUGG---AGGACUACAUACUAUGGUCCGUA--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ...(((.(((..((((.---.......))))...))).)))..--..(--((((.(((....))).))))).((((((((((........)))))))))).... ( -31.80, z-score = -2.33, R) >droSec1.super_3 3359056 97 - 7220098 ACUCGGUGUACAUAUGG---AGGACUACAUACUGUGGUCCGCA--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ....(((((.(....).---.(((((((.....))))))))))--))(--((((.(((....))).))))).((((((((((........)))))))))).... ( -38.60, z-score = -3.73, R) >droYak2.chr2L 17272187 80 + 22324452 ----------UCUAUGG---AGG-------ACCGUGGUCCGUG--CUG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ----------.......---.((-------((....))))..(--(((--((((.(((....))).))))).((((((((((........))))))))))))). ( -31.40, z-score = -3.25, R) >droEre2.scaffold_4929 16766346 86 - 26641161 ----------UCUAUGG---AGGACCAUGGUCUGU-ACUCGCA--CAG--UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ----------.((((((---....)))))).....-.......--..(--((((.(((....))).))))).((((((((((........)))))))))).... ( -30.00, z-score = -2.31, R) >droAna3.scaffold_12943 3579462 104 + 5039921 AACCGUAGUACAUUCUGCUCAUGUGUACCUUGUAUGGUUUGUAGUCUGGCUUGCUGGCUUCUCGCUGGAAUGUUAUUUGCCUCAGUUGUUGGGCAAAUAAAGUU .((((((((((((.........))))))....))))))((.((((..((((....))))....)))).))..((((((((((........)))))))))).... ( -26.20, z-score = -0.64, R) >consensus A_UCGGU_UACAUAUGG___AGGACUACAUACUGUGGUCCGUA__CUG__UUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUU ..................................................((((.(((....))).))))..((((((((((........)))))))))).... (-17.41 = -17.77 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:30 2011