
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,777,981 – 13,778,072 |
| Length | 91 |
| Max. P | 0.692212 |

| Location | 13,777,981 – 13,778,072 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Shannon entropy | 0.25779 |
| G+C content | 0.43701 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13777981 91 + 27905053 UUUUAAUCAAACUGCGACUUGGCCACGUUUUGCUUUAGCGCCUUUUUUUUUAAGGUGCGGUUCAGUGGCCUUU-GGUUGGUCAUAAAACCAA ..............(((((.((((((.....(((...(((((((.......))))))))))...))))))...-)))))............. ( -28.00, z-score = -2.34, R) >droAna3.scaffold_13340 11762407 90 - 23697760 UUUUAAUCAAACUGCGACUUGGCCACAGUUUACUUUAGCGCCUUUUUUU--AAGGUGCGGUUCAGUGGCCUUUUGGGUGGCCAUAAAACCAA ........((((((.(.......).))))))......(((((((.....--)))))))((((..((((((........))))))..)))).. ( -31.40, z-score = -2.98, R) >droEre2.scaffold_4770 9897328 88 - 17746568 UUUUAAUCAAACUGAGACUUGGCCACGUUUUGCUUUAGCGCCUUUUUU---AAGGUGCGGUUCAGUGGCCUUU-GGGUGGUCAUAAAACCAA ..........(((.(((...((((((.....(((...(((((((....---))))))))))...)))))))))-.)))(((......))).. ( -27.20, z-score = -1.88, R) >droYak2.chr3R 2370481 86 + 28832112 UUUUAAUCAAACUG--ACUUGGCCACGUUUUGCUUUAGCGCCGUUUUU---AAGGUGCGGUUCAGUGGCCUUU-GGGUGGUCAUAAAACCAA ...........(..--(...((((((.....(((...(((((......---..))))))))...)))))).).-.).((((......)))). ( -25.70, z-score = -1.48, R) >droSec1.super_12 1745422 89 - 2123299 UUUUAAUCAAACUGCGACUUGGCCACGUUUUGCUUUAGCGCCUUUUUUU--AAGGUGCGGUUCAGUGGCCUUU-GGGUGGUCAUAAAACCAA .............((((..((....))..))))....(((((((.....--)))))))((((..((((((...-....))))))..)))).. ( -27.80, z-score = -1.92, R) >droSim1.chr3R 19804336 88 - 27517382 UUUUAAUCAAACUGCGACUUGGCCACGUUUUGCUUUAGCGCCUUUUUU---AAGGUGCGGUUCAGUGGCCUUU-GGGUGGUCAUAAAACCAA .............((((..((....))..))))....(((((((....---)))))))((((..((((((...-....))))))..)))).. ( -27.80, z-score = -1.95, R) >droWil1.scaffold_181089 7459542 84 - 12369635 UUUUAAUCAAGCCG---CCUUGCUACAGUUGACUUUAGCGCCUUUUUU---AAGGUGCGGUUCAGUGCCCUUU--GGUGCCCAUAAAAGUGA (((((....(((((---(...((((.((....)).))))(((((....---)))))))))))..(..((....--))..)...))))).... ( -22.70, z-score = -1.17, R) >droMoj3.scaffold_6540 31363624 82 - 34148556 UUUUAAUCAAACCG---CC--GCUAGAGUUUACUUUUGCGCCUUUUUU---AAGGUGCGGUUCAGUGCCCUUU--GGUGGCCAUAAAACAGA .............(---((--(((((((..((((.(((((((((....---)))))))))...))))..))))--))))))........... ( -30.80, z-score = -4.22, R) >droGri2.scaffold_14830 3472937 83 - 6267026 UUUUAAUCAAACCG---CC--GCUACAGUUUACUUUUGCGCCUUUUUUU--AAGGUGCGGUUCAGUGCCCUUU--GGUGGCCAUAAAACUGA .............(---((--((((.((..((((.(((((((((.....--)))))))))...))))..)).)--))))))........... ( -27.20, z-score = -3.17, R) >droVir3.scaffold_13047 12304430 82 - 19223366 UUUUAAUCAAACCG---CC--GCUUCAGUUUACUUUUGCGCCUUUUUU---AAGGUGCGGUUCAGUGCCCUUU--GGUGGCCAUAAAACAGA .............(---((--(((..((..((((.(((((((((....---)))))))))...))))..))..--))))))........... ( -26.00, z-score = -2.87, R) >consensus UUUUAAUCAAACUG__ACUUGGCCACAGUUUACUUUAGCGCCUUUUUU___AAGGUGCGGUUCAGUGGCCUUU_GGGUGGCCAUAAAACCAA .....................(((((...........(((((((.......)))))))((........))......)))))........... (-15.61 = -15.62 + 0.01)
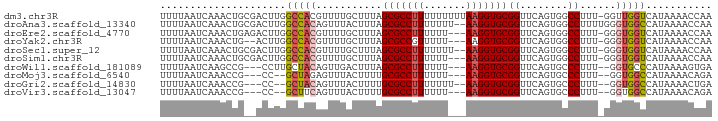
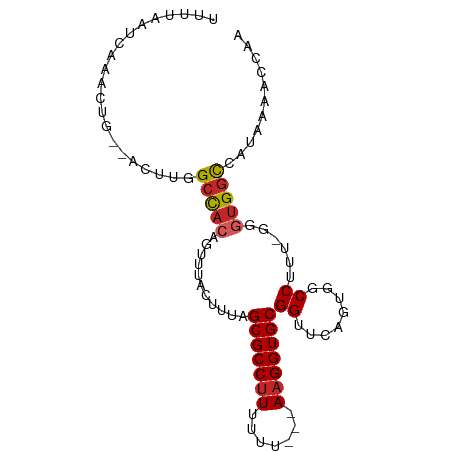

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:58 2011