| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,775,036 – 13,775,115 |
| Length | 79 |
| Max. P | 0.913082 |

| Location | 13,775,036 – 13,775,115 |
|---|---|
| Length | 79 |
| Sequences | 7 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.55586 |
| G+C content | 0.49132 |
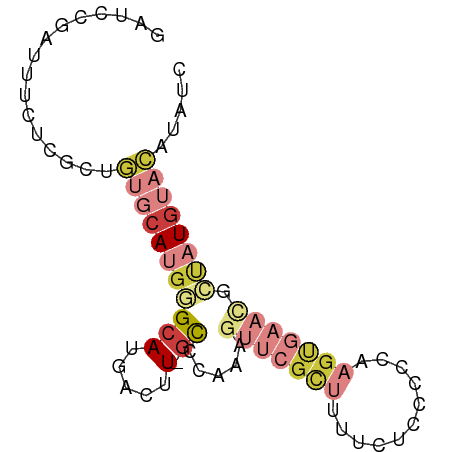
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -6.44 |
| Energy contribution | -8.20 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
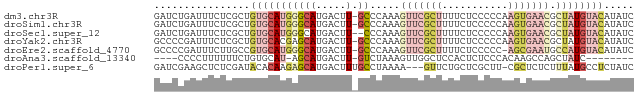
>dm3.chr3R 13775036 79 - 27905053 GAUCUGAUUUCUCGCUGUGCAUGGGCAUGACUU-GCCCAAAGUUCGCUUUUCUCCCCCAAGUGAACGCUAUGUACAUAUC ...............((((((((((((.....)-))))...((((((((.........))))))))...))))))).... ( -24.30, z-score = -3.56, R) >droSim1.chr3R 19801401 79 + 27517382 GAUCUGAUUUCUCGCUGUGCAUGGGCAUGACUU-GCCCAAAGUUCGCUUUUCUCCCCCAAGUGAACGCUAUGUACAUAUC ...............((((((((((((.....)-))))...((((((((.........))))))))...))))))).... ( -24.30, z-score = -3.56, R) >droSec1.super_12 1742519 78 + 2123299 GAUCUGAUUUCUCGCUGUGCAUGGGCAUGACUU--CCCAAAGUUCGCUUUUCUCCCCCAAGUGAACGCUAUGUACAUAUC ...............((((((((((........--)))...((((((((.........))))))))...))))))).... ( -18.20, z-score = -1.75, R) >droYak2.chr3R 2367451 79 - 28832112 GCCCCGAUUUCUCGCUGUGCACGAGCAUGACUU-GCCCAAAGUUCGCUUUUCUCCCCCAAGUGAACGCUAUGUACAUAUC ....(((....))).((((((.(.(((.....)-)).)...((((((((.........))))))))....)))))).... ( -16.70, z-score = -1.86, R) >droEre2.scaffold_4770 9894526 78 + 17746568 GCCCCGAUUUCUUGCCGUGCAUGGGCAUGACUU-GCCCAAAGUUCGCUUUUCUCCCCC-AGCGAAUGCCAUGUACAUAUC ................(((((((((((.....)-))))...(((((((..........-)))))))...))))))..... ( -22.80, z-score = -2.88, R) >droAna3.scaffold_13340 11759895 66 + 23697760 ----CCCCUUUUUUCUGUGCAU-AGCAUGACUU-GUCUAAAGUUGGCUCCACUCUCCCACAAGCCAGCUAUC-------- ----................((-(((..(((((-.....)))))((((.............)))).))))).-------- ( -11.72, z-score = -1.01, R) >droPer1.super_6 4121658 76 - 6141320 GAUCGAAGCUCUCGAUACACAAGAGCAUGACUUUGCCUAAAA---GUUCUGCUCGCUU-CGCUCUCUUUAUGCCUCUAUC ((.((((((....(.....)..(((((.((((((.....)))---))).)))))))))-)).))................ ( -21.50, z-score = -3.19, R) >consensus GAUCCGAUUUCUCGCUGUGCAUGGGCAUGACUU_GCCCAAAGUUCGCUUUUCUCCCCCAAGUGAACGCUAUGUACAUAUC ................((((((.(((.((........))..(((((((...........))))))))))))))))..... ( -6.44 = -8.20 + 1.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:55 2011