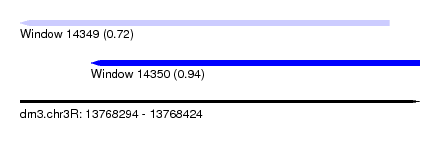
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,768,294 – 13,768,424 |
| Length | 130 |
| Max. P | 0.944892 |

| Location | 13,768,294 – 13,768,414 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.46796 |
| G+C content | 0.52965 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13768294 120 - 27905053 GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGUCGAAAGCCCCAAAACACGCUUCCUGGCCACCAAUGCACCAUGGCGGUAG ....(((((.((((((....(((((.....))))).((....((((((((...))))))))(((((.(((((....)...........))))..)))))......)).))))))))))). ( -41.30, z-score = -1.76, R) >droAna3.scaffold_13340 11753580 91 + 23697760 AGUGCACCGUCCAUGCCAGCGUUCAUUAAUUGAAGAGGUCG-AAGUGCAAAGCUAUUGCUUCCUCAAAAG------GGCACAAAAAAC-CCCAACAUCC--------------------- .((((.((....(((....))).......((((.((((.((-((((.....))).))))))).))))..)------))))).......-..........--------------------- ( -18.40, z-score = -0.45, R) >droEre2.scaffold_4770 9887772 99 + 17746568 GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCGCCGAAUGCUUGGCCA--------------CAAAACACGCUUCCCAGCCAGCA-------AUAGUGGUAU .(((((((((.....)))))(((((.....))))).(((....)))))))((((..((((.(((..--------------................)))))))-------....)))).. ( -30.07, z-score = -0.69, R) >droYak2.chr3R 2355487 119 - 28832112 GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACACAUCGAAAGCGCUCCCCAAAUGCUUGGCCACAAGU-GUAAGCCCCAAAACACGCUCCAUAGCCACCAAUACUCCAUAGUGGUAU ((((((.(((....(((((((((((.....)))))....(....).)))......(((((((....)))))-))..))).....)))))))))...(((((............))))).. ( -35.40, z-score = -2.81, R) >droSec1.super_12 1735937 119 + 2123299 GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGU-CUAAGCCCCAACACACGCUUCCUGGCCACCAAUGCACCAUGGCGGUAU ...((((((.((((((....(((((.....))))).((....((((((((...))))))))(((((.((((-................))))..)))))......)).)))))))))))) ( -40.99, z-score = -1.69, R) >droSim1.chr3R 19794488 119 + 27517382 GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGU-CUAAGCCCCAAAACACGCUUCCUGGCCACCAAUGCACCAUGGCGGUAU ...((((((.((((((....(((((.....))))).((....((((((((...))))))))(((((.((((-................))))..)))))......)).)))))))))))) ( -40.99, z-score = -1.84, R) >consensus GGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGU__UAAGCCCCAAAACACGCUUCCUAGCCACCAAUGCACCAUGGCGGUAU ((((((((.((...(((...(((((.....))))).))).)).))))))))((((....))))........................((((.....................)))).... (-18.14 = -18.72 + 0.58)

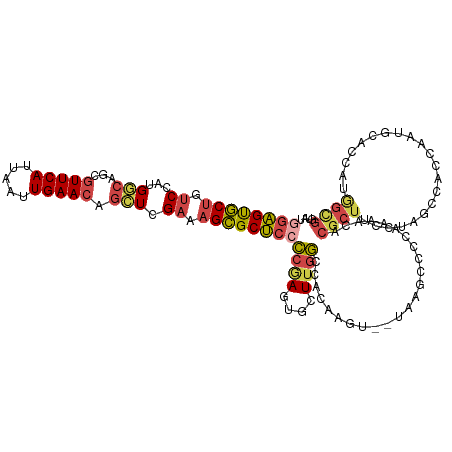
| Location | 13,768,317 – 13,768,424 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.46270 |
| G+C content | 0.53706 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -20.93 |
| Energy contribution | -20.72 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13768317 107 - 27905053 -------------CCACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGUCGAAAGCCCCAAAACACGCUUCCUGG -------------......((.(((((((.(((....(((.(((((((.....))))).))((((((((((((...))))))))(....)...))))..))).....)))))))))))). ( -35.50, z-score = -2.05, R) >droAna3.scaffold_13340 11753588 106 + 23697760 GUGCUGCCAAUUUGGGCGUCAUAAGUGCACCGUCCAUGCCAGCGUUCAUUAAUUGAAGAGGUCG-AAGUGCAAAGCUAUUGCUUCCUCAAAAG------GGCACAAAAAACCC------- .((.((((..((((((.........(((((((.((.((....))((((.....))))..)).))-..)))))((((....)))).))))))..------))))))........------- ( -26.30, z-score = -0.04, R) >droEre2.scaffold_4770 9887788 93 + 17746568 -------------CCACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCGCCGAAUGCUUGGCCACAAAACACGCUUCCCAG-------------- -------------........((((((((((((.....)))))(((((.....)))))(((.(....).))).(((((....)))))..........)).))))).-------------- ( -31.00, z-score = -2.21, R) >droYak2.chr3R 2355510 106 - 28832112 -------------CUACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACACAUCGAAAGCGCUCCCCAAAUGCUUGGCCACAAGUG-UAAGCCCCAAAACACGCUCCAUAG -------------..........((((((.(((....(((((((((((.....)))))....(....).)))......(((((((....))))))-)..))).....))))))))).... ( -30.50, z-score = -2.12, R) >droSec1.super_12 1735960 106 + 2123299 -------------CCACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGUC-UAAGCCCCAACACACGCUUCCUGG -------------......((.(((((((.(((....(((((((((((.....))))).)))...((((((((...))))))))(((.....)))-...))).....)))))))))))). ( -34.30, z-score = -1.70, R) >droSim1.chr3R 19794511 106 + 27517382 -------------CCACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGUC-UAAGCCCCAAAACACGCUUCCUGG -------------......((.(((((((.(((....(((((((((((.....))))).)))...((((((((...))))))))(((.....)))-...))).....)))))))))))). ( -34.30, z-score = -1.85, R) >consensus _____________CCACCACAUGGGAGUGCUGUCCAUGGCAGCGUUCAUUAAUUGAACAGCUCGAAAGCGCUCCCCGAGUGCUUGGCCACAAGUC_UAAGCCCCAAAACACGCUUCCUGG .............(((...((((((((((((.((...(((...(((((.....))))).))).)).)))))))))...)))..))).................................. (-20.93 = -20.72 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:53 2011