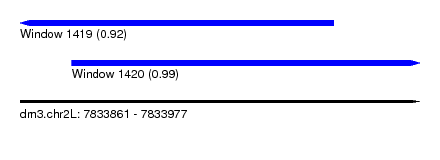
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,833,861 – 7,833,977 |
| Length | 116 |
| Max. P | 0.985426 |

| Location | 7,833,861 – 7,833,952 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Shannon entropy | 0.36911 |
| G+C content | 0.44498 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.72 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
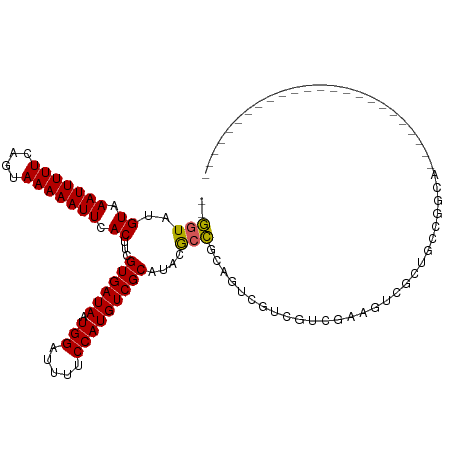
>dm3.chr2L 7833861 91 - 23011544 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGUCGUCGACGUCGCUGCCUGCA------------------------ --.((((((.(((((((....))))))).))...((((((.(((.....)))))))))))))((.((((.((.((....)).))))))..)).------------------------ ( -26.00, z-score = -2.25, R) >droSec1.super_3 3348044 91 - 7220098 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGUCGUCGACGUCGCUGCCUGCA------------------------ --.((((((.(((((((....))))))).))...((((((.(((.....)))))))))))))((.((((.((.((....)).))))))..)).------------------------ ( -26.00, z-score = -2.25, R) >droYak2.chr2L 17260813 91 + 22324452 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGUCGUCGACGUCGCUGCCAGCA------------------------ --.((((((.(((((((....))))))).))...((((((.(((.....)))))))))))))((.((((.((.((....)).))))))..)).------------------------ ( -26.00, z-score = -2.37, R) >droEre2.scaffold_4929 16755272 91 - 26641161 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGUCGUCGACGUCGCUGCCUGCA------------------------ --.((((((.(((((((....))))))).))...((((((.(((.....)))))))))))))((.((((.((.((....)).))))))..)).------------------------ ( -26.00, z-score = -2.25, R) >droAna3.scaffold_12943 3566177 115 + 5039921 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCACAGUCGCAGUCGCAGUAGCAGUCGCUGCCUCAGUCGACUUCGCUGCCAGAA --(((..((.(((((((....))))))).))...((((((.(((.....)))))))))....))).(((.((.((((((.(((((....)))))....).))))).)))))...... ( -31.30, z-score = -1.92, R) >dp4.chr4_group3 6227376 103 + 11692001 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGCAGUCGCAGUAGCAGUGGCUGUCGCUGCCCCAA------------ --(((..((.(((((((....))))))).))...((((((.(((.....)))))))))....)))(((((.((((((((.......)))))))).))))).....------------ ( -34.70, z-score = -3.82, R) >droPer1.super_1 3327079 103 + 10282868 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGCAGUCGCAGUAGCAGUGGCUGUCGCUGCCCCAA------------ --(((..((.(((((((....))))))).))...((((((.(((.....)))))))))....)))(((((.((((((((.......)))))))).))))).....------------ ( -34.70, z-score = -3.82, R) >droWil1.scaffold_180703 443070 88 + 3946847 --GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGAUGUCGACGUCGCUGCUA--------------------------- --(((..((.(((((((....))))))).))...((((((.(((.....)))))))))....)))((((.(((((....)))))))))..--------------------------- ( -27.60, z-score = -3.23, R) >droMoj3.scaffold_6500 29605633 87 + 32352404 --GCUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACAUCGCAGUAGCUGUCGCAGUCGCAGCC---------------------------- --((.((((((((((((....)))))))......((((((.(((.....))))))))).))))).))....((((.((....)))))).---------------------------- ( -22.00, z-score = -2.51, R) >droGri2.scaffold_15252 3994716 88 + 17193109 GGUAUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAU-CGCAGUAGCUGUCGCCCCACUCACCCUC---------------------------- (((....((.(((((((....))))))).))...((((((.(((.....)))))))))..-.((((...)))).)))............---------------------------- ( -14.60, z-score = -0.78, R) >consensus __GGUAUGUAAAUUUUUCAGUAAAAAUUCACUUCGUGAUAAUGGAUUUUCCAUGUCGCAUACGCCGCAGUCGUCGUCGAAGUCGCUGCCGGCA________________________ ..(((..((.(((((((....))))))).))...((((((.(((.....)))))))))....))).................................................... (-13.79 = -13.72 + -0.07)

| Location | 7,833,876 – 7,833,977 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.52841 |
| G+C content | 0.47986 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
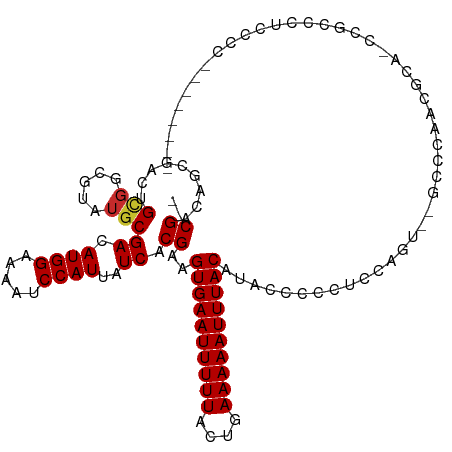
>dm3.chr2L 7833876 101 + 23011544 --CGACGACGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCUCUCCAGU-AACCCAACGCA-CCGCA-------------- --...........((((....((((..((((.....)))).........((((((((((....))))))))))..............-.......))))-)))).-------------- ( -21.50, z-score = -1.87, R) >droSec1.super_3 3348059 107 + 7220098 --CGACGACGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCUCUCCAGUGCCCCCAACGCA-CCGCACCGCA--------- --...........((((.((.((((..((((.....)))).........((((((((((....))))))))))......................))))-..)).)))).--------- ( -25.30, z-score = -2.24, R) >droAna3.scaffold_12943 3566214 100 - 5039921 UGCGACUGCGACUGUGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCACUCCGCU---ACUGGCGCA-GCGC--------------- .(((.(((((...((((((..((.((.((((.....))))..)).))..((((((((((....))))))))))..........))))---))...))))-))))--------------- ( -33.40, z-score = -4.23, R) >dp4.chr4_group3 6227401 106 - 11692001 UGCGACUGCGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCCCACA-----GCUCUCAUCUCUCCCCUUCCCC-------- ...((.((.(((((.((.(((((.((.((((.....))))..)).)...((((((((((....))))))))))))))..)).))-----).)).)))).............-------- ( -20.50, z-score = -1.86, R) >droPer1.super_1 3327104 106 - 10282868 UGCGACUGCGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCCCACA-----GCUCUCAUCUCUCCCCUUCCCC-------- ...((.((.(((((.((.(((((.((.((((.....))))..)).)...((((((((((....))))))))))))))..)).))-----).)).)))).............-------- ( -20.50, z-score = -1.86, R) >droWil1.scaffold_180703 443082 113 - 3946847 --CGACAUCGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCAUUCCCUUCAAACCAACACAACCGCCCCCCCGCGUU---- --.......(((.((((.(...(((..((((.....)))).........((((((((((....))))))))))............................)))...))))))))---- ( -21.00, z-score = -1.75, R) >droVir3.scaffold_12963 8599279 113 + 20206255 --CGACAGCUACUGCGAUGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUAG--CCGCGAGC--GCCCCCAACCCCCUUCGGCUCCCCGGACCC --.....(((...(((.(((.((.((.((((.....))))..)).))..((((((((((....))))))))))))).--.))).)))--.............(((((....)))))... ( -22.30, z-score = -0.72, R) >droMoj3.scaffold_6500 29605644 110 - 32352404 --CGACAGCUACUGCGAUGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUAG--CCGCGAGC--GUCCC--GCCUCCUCCCUCCCGCCUGUCC- --.((((((....))(((((.((((.(((((.....)))).........((((((((((....))))))))))...)--.)))).))--)))..--.................)))).- ( -26.40, z-score = -2.50, R) >droGri2.scaffold_15252 3994730 106 - 17193109 --CGACAGCUACUGCGAUG----CGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUAUACCCGCGAGCUGACUGCUGACUGCCCCCCUCCCCC------- --...((((....((..((----((..((((.....)))).........((((((((((....)))))))))).......)))).)).....))))................------- ( -23.20, z-score = -4.01, R) >consensus __CGACAGCGACUGCGGCGUAUGCGACAUGGAAAAUCCAUUAUCACGAAGUGAAUUUUUACUGAAAAAUUUACAUACCCCCUCCAGU__GCCCAACGCA_CCGCCCUCCCC________ ..((.........(((.....)))((.((((.....))))..)).))..((((((((((....)))))))))).............................................. (-11.46 = -11.60 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:27 2011