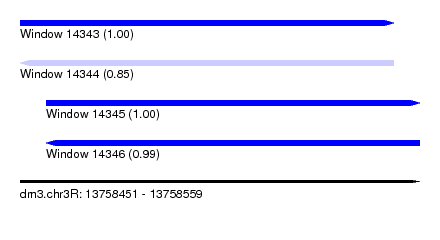
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,758,451 – 13,758,559 |
| Length | 108 |
| Max. P | 0.997908 |

| Location | 13,758,451 – 13,758,552 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.57617 |
| G+C content | 0.31067 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -13.49 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13758451 101 + 27905053 AUUAAUAGGCAUUGGGUGCAUAUAUAUAAAAUUUGCAUAGGUACCUAAAAGUAUGCAACAAUAUAUAGUUGGCC------------------CAUUUUUUGUUGCAUACUUUUACACAC .....((((.(((..(((((.............))))).))).))))((((((((((((((.............------------------......))))))))))))))....... ( -25.53, z-score = -1.94, R) >droSim1.chr3R 19784755 118 - 27517382 AUCAAAAAGCAUUAAUUGCAUAUUUAUAUCAUUUGUAUAGAUAUCUGAACGUAUGCAACAAUACAU-UCAAGCCAAUUUGAGAGAAACACACACUUUUUCGUUGCAUACUUUUACACAC .(((....(((.....)))((((((((((.....)))))))))).)))..(((((((((.......-(((((....)))))((((((.......))))))))))))))).......... ( -26.10, z-score = -4.44, R) >droSec1.super_12 1726139 112 - 2123299 ------AUAAAAAAAUUGCAUAUUUAUAUAAUUUGUAUAGAUAUUUAAACGUAUUCAACCAUACAU-UUAAGCCAAUUUGAGAGGCACACACACUUUUUCGUUGAAUACUUUUACACAC ------.(((((...(((.((((((((((.....)))))))))).)))..(((((((((.......-....(((.........)))..............))))))))))))))..... ( -17.71, z-score = -2.75, R) >droYak2.chr3R 2345318 118 + 28832112 AUAAGCAAGCAUUAAUUGGCUAUUUAUAUCAUGUCUAUGGGUAGCUAAAAGUAUGCAACAAUAUAU-GUAAGCCGUUUGAAUAGACCCACACGCUCUGUUGUUGCAUACUUUUACACAU .................(((((((((((.......)))))))))))(((((((((((((((((...-...(((.((.((........)).))))).)))))))))))))))))...... ( -31.90, z-score = -2.44, R) >droEre2.scaffold_4770 9878277 99 - 17746568 AUUAAAAAGCGUUAACUGCAUAUUUUUAUCAUUUCUGUGGGGACCUAAAAGUAAGCAACAAGAAAU-GUAAGC-------------------GCUAUUUUGCUGCAUACUUUUGCACAC .......((((((...(((.((((((((...(..(....)..)..)))))))).)))(((.....)-)).)))-------------------)))....((.((((......)))))). ( -19.00, z-score = -0.08, R) >consensus AUUAAAAAGCAUUAAUUGCAUAUUUAUAUCAUUUGUAUAGGUACCUAAAAGUAUGCAACAAUACAU_GUAAGCC__UU__A_AG___CACACACUUUUUUGUUGCAUACUUUUACACAC ...................(((((((((.......))))))))).((((((((((((((((.....................................))))))))))))))))..... (-13.49 = -14.45 + 0.96)

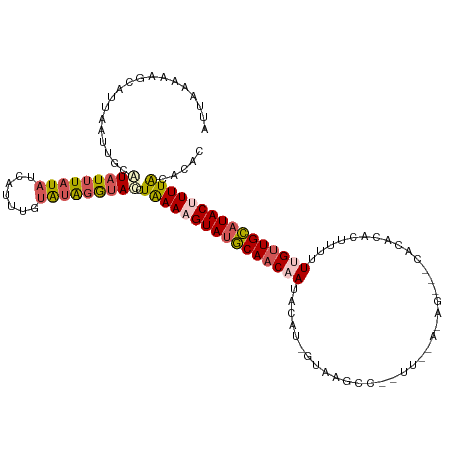
| Location | 13,758,451 – 13,758,552 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.57617 |
| G+C content | 0.31067 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -13.67 |
| Energy contribution | -14.11 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13758451 101 - 27905053 GUGUGUAAAAGUAUGCAACAAAAAAUG------------------GGCCAACUAUAUAUUGUUGCAUACUUUUAGGUACCUAUGCAAAUUUUAUAUAUAUGCACCCAAUGCCUAUUAAU .....((((((((((((((((.(.(((------------------(.....)))).).))))))))))))))))((((....((((.((.......)).)))).....))))....... ( -28.30, z-score = -3.03, R) >droSim1.chr3R 19784755 118 + 27517382 GUGUGUAAAAGUAUGCAACGAAAAAGUGUGUGUUUCUCUCAAAUUGGCUUGA-AUGUAUUGUUGCAUACGUUCAGAUAUCUAUACAAAUGAUAUAAAUAUGCAAUUAAUGCUUUUUGAU ((((((....(((((((((((((.........))))..((((......))))-.......)))))))))......(((((.........)))))..))))))................. ( -20.50, z-score = 0.42, R) >droSec1.super_12 1726139 112 + 2123299 GUGUGUAAAAGUAUUCAACGAAAAAGUGUGUGUGCCUCUCAAAUUGGCUUAA-AUGUAUGGUUGAAUACGUUUAAAUAUCUAUACAAAUUAUAUAAAUAUGCAAUUUUUUUAU------ ...................(((((((((((((((((.........)))((((-((((((......)))))))))).....................)))))).))))))))..------ ( -18.90, z-score = -0.97, R) >droYak2.chr3R 2345318 118 - 28832112 AUGUGUAAAAGUAUGCAACAACAGAGCGUGUGGGUCUAUUCAAACGGCUUAC-AUAUAUUGUUGCAUACUUUUAGCUACCCAUAGACAUGAUAUAAAUAGCCAAUUAAUGCUUGCUUAU .....((((((((((((((((...(..(((((((((.........)))))))-)).).))))))))))))))))((((...(((.......)))...)))).................. ( -30.10, z-score = -1.65, R) >droEre2.scaffold_4770 9878277 99 + 17746568 GUGUGCAAAAGUAUGCAGCAAAAUAGC-------------------GCUUAC-AUUUCUUGUUGCUUACUUUUAGGUCCCCACAGAAAUGAUAAAAAUAUGCAGUUAACGCUUUUUAAU (((((((((((((.(((((((......-------------------......-.....))))))).))))))).........((....)).............))..))))........ ( -17.23, z-score = 0.39, R) >consensus GUGUGUAAAAGUAUGCAACAAAAAAGCGUGUG___CU_U__AA__GGCUUAC_AUAUAUUGUUGCAUACUUUUAGAUACCUAUACAAAUGAUAUAAAUAUGCAAUUAAUGCUUUUUAAU (((((((((((((((((((((.....................................))))))))))))))))......))))).................................. (-13.67 = -14.11 + 0.44)

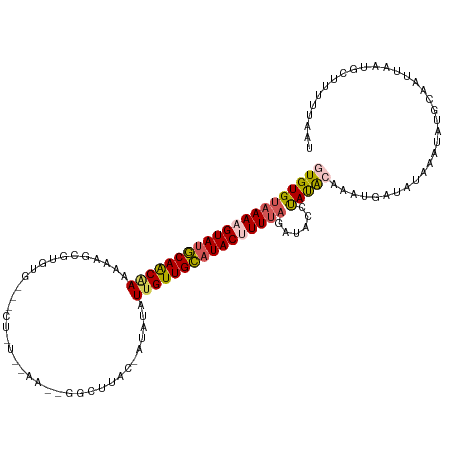
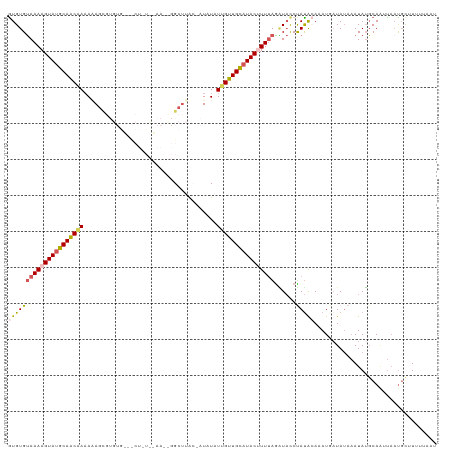
| Location | 13,758,458 – 13,758,559 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.56138 |
| G+C content | 0.33486 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13758458 101 + 27905053 GGCAUUGGGUGCAUAUAUAUAAAAUUUGCAUAGGUACCUAAAAGUAUGCAACAAUAUAU-------AGUU-----GGCCCA-----UUUUUUGUUGCAUACUUUUACACACCUCUCGA ....(((((((((.............)))).((((...((((((((((((((((.....-------....-----......-----....))))))))))))))))...))))))))) ( -26.73, z-score = -2.36, R) >droSim1.chr3R 19784762 118 - 27517382 AGCAUUAAUUGCAUAUUUAUAUCAUUUGUAUAGAUAUCUGAACGUAUGCAACAAUACAUUCAAGCCAAUUUGAGAGAAACACACACUUUUUCGUUGCAUACUUUUACACACCUCUCGA .(((.....)))((((((((((.....)))))))))).((((.(((((((((.......(((((....)))))((((((.......))))))))))))))).))))............ ( -25.90, z-score = -4.17, R) >droSec1.super_12 1726146 112 - 2123299 ------AAUUGCAUAUUUAUAUAAUUUGUAUAGAUAUUUAAACGUAUUCAACCAUACAUUUAAGCCAAUUUGAGAGGCACACACACUUUUUCGUUGAAUACUUUUACACACCUCUCGA ------....((((((((((((.....))))))))))(((((.((((......)))).)))))))....((((((((............(((...)))............)))))))) ( -18.35, z-score = -2.97, R) >droYak2.chr3R 2345325 118 + 28832112 AGCAUUAAUUGGCUAUUUAUAUCAUGUCUAUGGGUAGCUAAAAGUAUGCAACAAUAUAUGUAAGCCGUUUGAAUAGACCCACACGCUCUGUUGUUGCAUACUUUUACACAUCUUUAAG ..........(((((((((((.......)))))))))))(((((((((((((((((......(((.((.((........)).))))).)))))))))))))))))............. ( -31.90, z-score = -2.96, R) >droEre2.scaffold_4770 9878284 99 - 17746568 AGCGUUAACUGCAUAUUUUUAUCAUUUCUGUGGGGACCUAAAAGUAAGCAACAAGAAAUGUAAGC-------------------GCUAUUUUGCUGCAUACUUUUGCACACCUCUAAA ((((((...(((.((((((((...(..(....)..)..)))))))).)))(((.....))).)))-------------------)))....((.((((......))))))........ ( -18.70, z-score = 0.17, R) >consensus AGCAUUAAUUGCAUAUUUAUAUCAUUUGUAUAGGUACCUAAAAGUAUGCAACAAUACAUGUAAGCCAAUU__A_AGACACACACACUUUUUUGUUGCAUACUUUUACACACCUCUCGA ............(((((((((.......))))))))).((((((((((((((((....................................))))))))))))))))............ (-13.52 = -14.48 + 0.96)

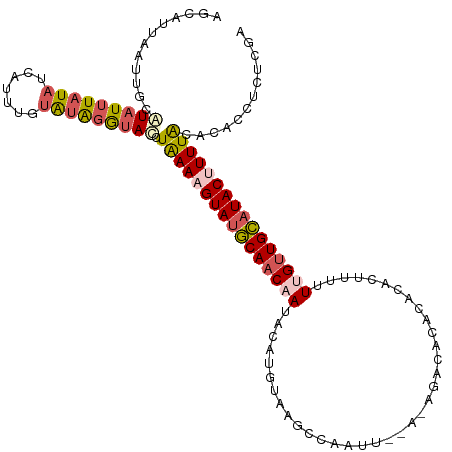
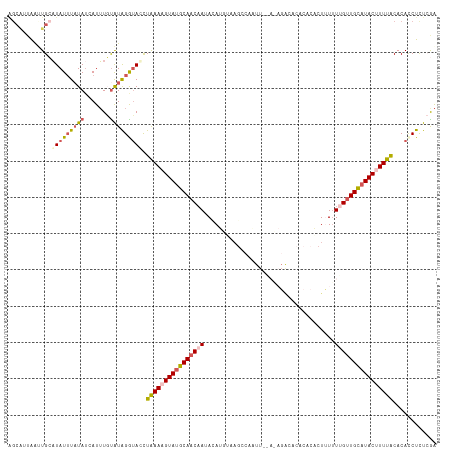
| Location | 13,758,458 – 13,758,559 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.56138 |
| G+C content | 0.33486 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13758458 101 - 27905053 UCGAGAGGUGUGUAAAAGUAUGCAACAAAAAA-----UGGGCC-----AACU-------AUAUAUUGUUGCAUACUUUUAGGUACCUAUGCAAAUUUUAUAUAUAUGCACCCAAUGCC .....(((((..((((((((((((((((.(.(-----(((...-----..))-------)).).))))))))))))))))..))))).((((.((.......)).))))......... ( -32.70, z-score = -4.19, R) >droSim1.chr3R 19784762 118 + 27517382 UCGAGAGGUGUGUAAAAGUAUGCAACGAAAAAGUGUGUGUUUCUCUCAAAUUGGCUUGAAUGUAUUGUUGCAUACGUUCAGAUAUCUAUACAAAUGAUAUAAAUAUGCAAUUAAUGCU ..((((((((((((....))))))((......)).......)))))).(((((..((((((((((......))))))))))(((((.........))))).......)))))...... ( -22.90, z-score = -0.20, R) >droSec1.super_12 1726146 112 + 2123299 UCGAGAGGUGUGUAAAAGUAUUCAACGAAAAAGUGUGUGUGCCUCUCAAAUUGGCUUAAAUGUAUGGUUGAAUACGUUUAAAUAUCUAUACAAAUUAUAUAAAUAUGCAAUU------ ..((((((((..((......(((...)))......))..))))))))......((((((((((((......))))))))))((((.((((.......)))).))))))....------ ( -23.90, z-score = -2.41, R) >droYak2.chr3R 2345325 118 - 28832112 CUUAAAGAUGUGUAAAAGUAUGCAACAACAGAGCGUGUGGGUCUAUUCAAACGGCUUACAUAUAUUGUUGCAUACUUUUAGCUACCCAUAGACAUGAUAUAAAUAGCCAAUUAAUGCU ............((((((((((((((((...(..(((((((((.........))))))))).).))))))))))))))))((((...(((.......)))...))))........... ( -30.10, z-score = -2.18, R) >droEre2.scaffold_4770 9878284 99 + 17746568 UUUAGAGGUGUGCAAAAGUAUGCAGCAAAAUAGC-------------------GCUUACAUUUCUUGUUGCUUACUUUUAGGUCCCCACAGAAAUGAUAAAAAUAUGCAGUUAACGCU ......((((((((((((((.(((((((......-------------------...........))))))).))))))).........((....)).............))..))))) ( -17.93, z-score = 0.66, R) >consensus UCGAGAGGUGUGUAAAAGUAUGCAACAAAAAAGCGUGUGGGCCU_U__AAUUGGCUUAAAUAUAUUGUUGCAUACUUUUAGAUACCUAUACAAAUGAUAUAAAUAUGCAAUUAAUGCU ......(((((.((((((((((((((((....................................)))))))))))))))).)))))................................ (-16.44 = -17.32 + 0.88)

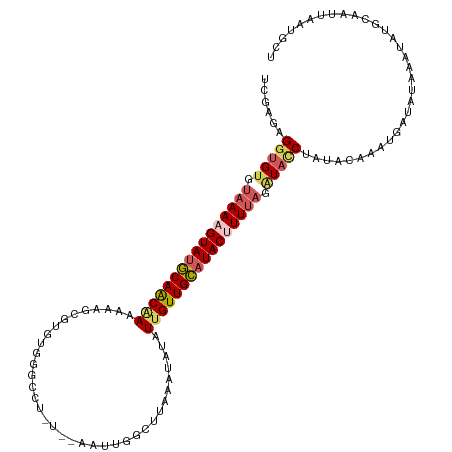
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:50 2011