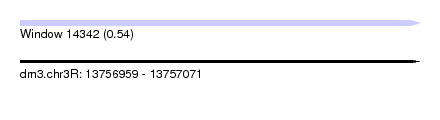
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,756,959 – 13,757,071 |
| Length | 112 |
| Max. P | 0.536393 |

| Location | 13,756,959 – 13,757,071 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 77.37 |
| Shannon entropy | 0.43022 |
| G+C content | 0.43143 |
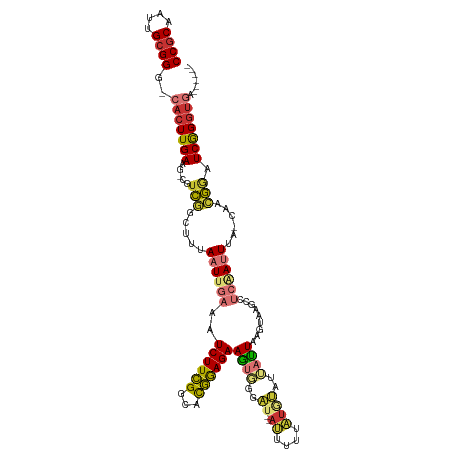
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.51 |
| Covariance contribution | -0.09 |
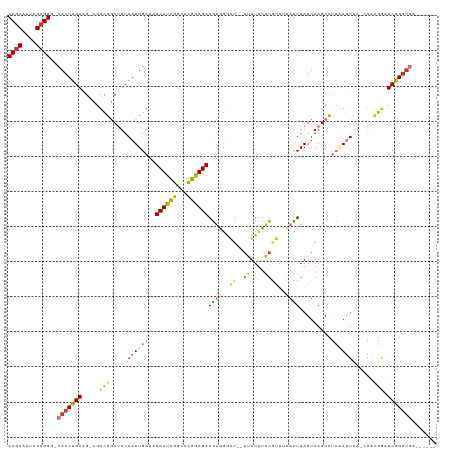
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
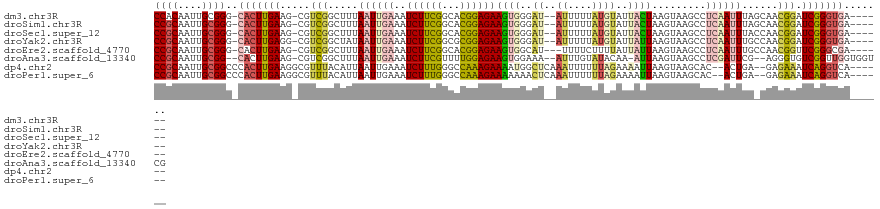
>dm3.chr3R 13756959 112 + 27905053 CCACAAUUGCGGG-CACUUGAAG-CGUCGGCUUUAAUUGAAAUCUUCGGCACGGAGAAGUGGGAU--AUUUUUAUGUAUUACUAAGUAAGCCUCAAUUUAGCAACGGAUCGGGUGA------ ((.(....).)).-(((((((..-(((..(((..((((((..((((((...))))))(((((.((--((....)))).))))).........)))))).))).)))..))))))).------ ( -27.50, z-score = -0.50, R) >droSim1.chr3R 19783239 112 - 27517382 CCGCAAUUGCGGG-CACUUGAAG-CGUCGGCUUUAAUUGAAAUCUUCGGCACGGAGAAGUGGGAU--AUUUUUAUGUAUUACUAAGUAAGCCUCAAUUUAGCAACGGAUCGGGUGA------ ((((....)))).-(((((((..-(((..(((..((((((..((((((...))))))(((((.((--((....)))).))))).........)))))).))).)))..))))))).------ ( -32.90, z-score = -1.78, R) >droSec1.super_12 1724615 112 - 2123299 CCGCAAUUGCGGG-CACUUGAAG-CGUCGGCUUUAAUUGAAAUCUUCGGCACGGAGAAGUGGGAU--AUUUUUAUGUAUUACUAAGUAAGCCUCAAUUUACCAACGGAUCGGGUGA------ ((((....)))).-(((((((..-(((.((....((((((..((((((...))))))(((((.((--((....)))).))))).........))))))..)).)))..))))))).------ ( -31.70, z-score = -1.42, R) >droYak2.chr3R 2343699 112 + 28832112 CCGCAAUUGCGGG-CACUUGAGG-CGUCGGCUAUAAUUGAAAUCUUCGGCGCGGAGAAGUGGGAU--AUUUUUAUGUAUUAUUAAGUAAGCCUCAAUUUGCCAACGGAUCGGGUGA------ ((((....)))).-(((((((..-(((.(((...((((((..((((((...)))))).....(((--((......)))))............)))))).))).)))..))))))).------ ( -35.20, z-score = -1.65, R) >droEre2.scaffold_4770 9876790 111 - 17746568 CCGCAAUUGCGGG-CACUUGAAG-CGUCGGCUUUAAUUGAAAUCUUCGGCACGGAGAAGUGGCAU---UUUUCUUUUAUUAUUAAGUAAGCCUCAAUUUGCCAACGGUUCGGGCGA------ ((((....)))).-(.((((((.-(((.(((...((((((..((((((...))))))...(((.(---(...(((........))).))))))))))).))).))).)))))).).------ ( -35.90, z-score = -2.12, R) >droAna3.scaffold_13340 11743016 114 - 23697760 CCGCAAUUGCGG--CACUUGAAG-CGUCGGCUUUAAUUGAAAUCUUCGUUUUGGAGAAGUGGAAA--AUUUGUAUACAA-AUUAAGUAAGCCUCGAUUCG--AGGGUGUCGGUUGGUGGUCG ((((....))))--((((.((((-(....)))))((((((.(((((((..(((..(........(--(((((....)))-))).......)..)))..))--))))).)))))))))).... ( -31.96, z-score = -1.25, R) >dp4.chr2 28770604 112 + 30794189 CCGCAAUUGCGGCCCACUUGAAGGCGUUUACAUUAAUUGAAAUCUUUGGGCCAAAGAAAAUGGCUCAAAUUUUUUAGAAAAUUAAGUAAGCAC--ACUGA--GAGAAAUCAGGUCA------ ((((....))))..........(((((((((.((((((((((..(((((((((.......)))))))))..)))))....)))))))))))..--.((((--......))))))).------ ( -33.60, z-score = -3.44, R) >droPer1.super_6 4096698 112 + 6141320 CCGCAAUUGCGGCCCACUUGAAGGCGUUUACAUUAAUUGAAAUCUUUGGGCCAAAGAAAAAAACUCAAAUUUUUUAGAAAAUUAAGUAAGCAC--ACUGA--GAGAAAUCAGGUCA------ ((((....))))..........(((((((((.((((((((((..((((((.............))))))..)))))....)))))))))))..--.((((--......))))))).------ ( -24.72, z-score = -1.40, R) >consensus CCGCAAUUGCGGG_CACUUGAAG_CGUCGGCUUUAAUUGAAAUCUUCGGCACGGAGAAGUGGGAU__AUUUUUAUGUAUUAUUAAGUAAGCCUCAAUUUA_CAACGGAUCGGGUGA______ ((((....))))..(((((((.....(((.....((((((..((((((...)))))).((((((.....)))))).................))))))......))).)))))))....... (-15.60 = -15.51 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:46 2011