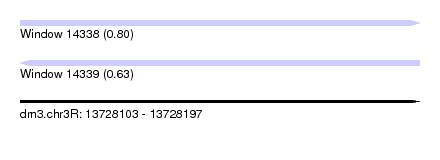
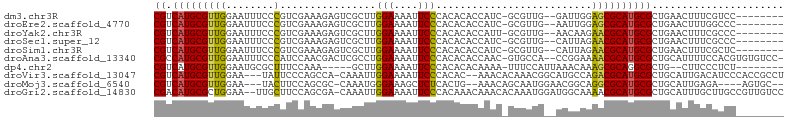
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,728,103 – 13,728,197 |
| Length | 94 |
| Max. P | 0.802441 |

| Location | 13,728,103 – 13,728,197 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.97 |
| Shannon entropy | 0.68862 |
| G+C content | 0.51659 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
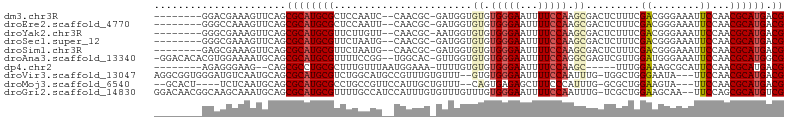
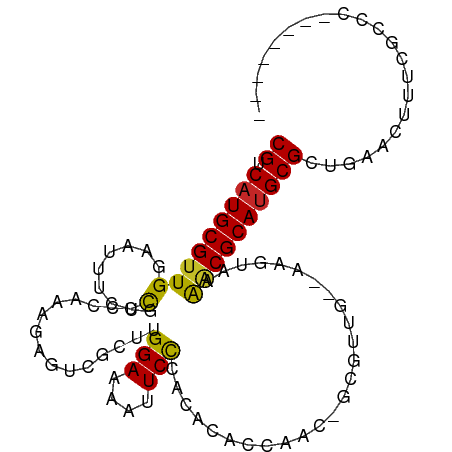
>dm3.chr3R 13728103 94 + 27905053 --------GGACGAAAGUUCAGCGCAUGCGCUCCAAUC--CAACGC-GAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG --------((((....))))..(((((((((.(((.((--......-))))).))((.((((((((((...(((.....)))..)))))))))).))))))).)) ( -35.10, z-score = -2.69, R) >droEre2.scaffold_4770 9849338 94 - 17746568 --------GGGCCAAAGUUCAGCGCAUGCGCUCCAAUU--CAACGC-GAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG --------((((....))))((((....))))......--.....(-(.(.((((((.((((((((((...(((.....)))..)))))))))).)))))).))) ( -29.90, z-score = -1.00, R) >droYak2.chr3R 2314588 94 + 28832112 --------GGGCGAAAGUUCAGCGCAUGCGUUCUUGUU--CAACGC-AAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG --------((((....))))..((((((((((......--..((((-(....))))).((((((((((...(((.....)))..)))))))))))))))))).)) ( -36.40, z-score = -3.16, R) >droSec1.super_12 1696415 94 - 2123299 --------GGGCGAAAGUUCAGCGCAUGCGUUCUAAUG--CAACGC-GAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG --------((((....)))).(((..(((((....)))--)).)))-(.(.((((((.((((((((((...(((.....)))..)))))))))).)))))).)). ( -36.70, z-score = -3.06, R) >droSim1.chr3R 19756493 94 - 27517382 --------GAGCGAAAGUUCAGCGCAUGCGUUCUAAUG--CAACGC-GAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG --------((((....)))).(((..(((((....)))--)).)))-(.(.((((((.((((((((((...(((.....)))..)))))))))).)))))).)). ( -37.70, z-score = -3.53, R) >droAna3.scaffold_13340 11712373 101 - 23697760 -GGACACACGUGGAAAAUGCAGCGCAUGCGUUUUCCGG--UGGCAC-GUUGGUGUGUGGGAAUUUUCCAGGCGAGUCGUUGGAUGGGAAAUUCCAACGCAUGGCG -.....(((.(((((((((((.....))))))))))))--))...(-(.(.((((((.(((((((((((..(((....)))..))))))))))).)))))).))) ( -43.00, z-score = -3.19, R) >dp4.chr2 28727141 89 + 30794189 --------AGAGGGAAG--CAGCGCCUGCGCUUUGUUUAAUGGAAA-UUUUGUGUGUGGGAAUUUUCCAAGC-----UUUGGAAAGCGCAUUCCAACGCAUGACG --------.(((.((((--(.((....))))))).)))........-..(..(((((.(((((((((((...-----..))))))....))))).)))))..).. ( -25.20, z-score = -0.32, R) >droVir3.scaffold_13047 12240636 99 - 19223366 AGGCGGUGGGAUGUCAAUGCAGCGCAUGCGUCUGGCAUGCCGUUUGUGUUU--GUGUGGGAAUUUUCCAAUUUG-UGGCUGGGAAUA---UUCCAACGCAUGACG .((((((.((((((..((((...)))))))))).)).))))......((((--((((.(((((((((((.....-....)))))).)---)))).))))).))). ( -35.20, z-score = -1.21, R) >droMoj3.scaffold_6540 31280949 93 - 34148556 --GCACU----UCUCAAUGCAGCGCAUGCGCCUGCCGUUCCAUUGCUGUUU--CAGUGAGAGCUUUCCCAUUUG-GCGCUGGAAGUA---UUCCAACGCAUGACG --.....----..(((..((((((....)).)))).(((((((((......--))))).))))...........-(((.((((....---.)))).))).))).. ( -24.10, z-score = 0.49, R) >droGri2.scaffold_14830 3410316 102 - 6267026 GGACAACGGCAAGCAAAUGCAGCGCAUGCGUUUUGCCAUCCAUUUGUGUUUGUUUGUGGGAAUUUUCCAAUUUG-UCGCUGGAAGCAA--UUCCAGCGCAUGUCG .((((.(.((((((((((((((.(.(((.(.....)))).)..)))))))))))))).).............((-.((((((((....--)))))))))))))). ( -36.80, z-score = -2.24, R) >consensus ________GGACGAAAGUUCAGCGCAUGCGUUCUAAUU__CAACGC_GAUGGUGUGUGGGAAUUUUCCAAGCGACUCUUUCGACGGGAAAUUCCAACGCAUGACG ....................((((....))))...................((((((.((((.........(((.....)))........)))).)))))).... ( -9.56 = -9.75 + 0.19)
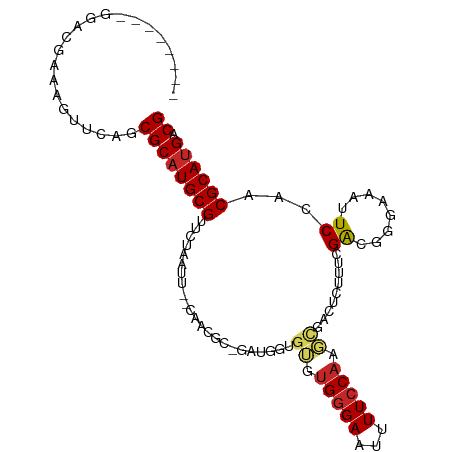
| Location | 13,728,103 – 13,728,197 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.97 |
| Shannon entropy | 0.68862 |
| G+C content | 0.51659 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -11.32 |
| Energy contribution | -10.91 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13728103 94 - 27905053 CGUCAUGCGUUGGAAUUUCCCGUCGAAAGAGUCGCUUGGAAAAUUCCCACACACCAUC-GCGUUG--GAUUGGAGCGCAUGCGCUGAACUUUCGUCC-------- ((.(((((((((((((((.(((.(((.....)))..))).)))))))......(((((-......--)).)))))))))))))..............-------- ( -26.80, z-score = -0.13, R) >droEre2.scaffold_4770 9849338 94 + 17746568 CGUCAUGCGUUGGAAUUUCCCGUCGAAAGAGUCGCUUGGAAAAUUCCCACACACCAUC-GCGUUG--AAUUGGAGCGCAUGCGCUGAACUUUGGCCC-------- .(.(((((((((((((((.(((.(((.....)))..))).)))))))......(((..-......--...))))))))))))((..(...)..))..-------- ( -26.40, z-score = -0.15, R) >droYak2.chr3R 2314588 94 - 28832112 CGUCAUGCGUUGGAAUUUCCCGUCGAAAGAGUCGCUUGGAAAAUUCCCACACACCAUU-GCGUUG--AACAAGAACGCAUGCGCUGAACUUUCGCCC-------- ..((((((((.(((((((.(((.(((.....)))..))).))))))).)).......(-(((((.--......)))))).))).)))..........-------- ( -24.20, z-score = -0.63, R) >droSec1.super_12 1696415 94 + 2123299 CGUCAUGCGUUGGAAUUUCCCGUCGAAAGAGUCGCUUGGAAAAUUCCCACACACCAUC-GCGUUG--CAUUAGAACGCAUGCGCUGAACUUUCGCCC-------- ..((((((((.(((((((.(((.(((.....)))..))).))))))).))........-(((((.--......)))))..))).)))..........-------- ( -23.80, z-score = -0.32, R) >droSim1.chr3R 19756493 94 + 27517382 CGUCAUGCGUUGGAAUUUCCCGUCGAAAGAGUCGCUUGGAAAAUUCCCACACACCAUC-GCGUUG--CAUUAGAACGCAUGCGCUGAACUUUCGCUC-------- ......((((.(((((((.(((.(((.....)))..))).))))))).))....((.(-((((.(--(........))))))).)).......))..-------- ( -24.20, z-score = -0.31, R) >droAna3.scaffold_13340 11712373 101 + 23697760 CGCCAUGCGUUGGAAUUUCCCAUCCAACGACUCGCCUGGAAAAUUCCCACACACCAAC-GUGCCA--CCGGAAAACGCAUGCGCUGCAUUUUCCACGUGUGUCC- ........((((((........))))))(((.(((.(((((((((((..(((......-)))...--..))))...(((.....))).))))))).))).))).- ( -28.80, z-score = -1.83, R) >dp4.chr2 28727141 89 - 30794189 CGUCAUGCGUUGGAAUGCGCUUUCCAAA-----GCUUGGAAAAUUCCCACACACAAAA-UUUCCAUUAAACAAAGCGCAGGCGCUG--CUUCCCUCU-------- .(.((.(((((....((((((((.....-----...((((((................-))))))......)))))))))))))))--)........-------- ( -19.23, z-score = -0.45, R) >droVir3.scaffold_13047 12240636 99 + 19223366 CGUCAUGCGUUGGAA---UAUUCCCAGCCA-CAAAUUGGAAAAUUCCCACAC--AAACACAAACGGCAUGCCAGACGCAUGCGCUGCAUUGACAUCCCACCGCCU .(((((((((.((((---(.((((.(....-....).)))).))))).))..--...........((((((.....))))))...))).))))............ ( -27.80, z-score = -2.52, R) >droMoj3.scaffold_6540 31280949 93 + 34148556 CGUCAUGCGUUGGAA---UACUUCCAGCGC-CAAAUGGGAAAGCUCUCACUG--AAACAGCAAUGGAACGGCAGGCGCAUGCGCUGCAUUGAGA----AGUGC-- ((.(((((((((((.---....))))))((-(...(((((....)))))(((--...((....))...)))..))))))))))..(((((....----)))))-- ( -31.30, z-score = -1.06, R) >droGri2.scaffold_14830 3410316 102 + 6267026 CGACAUGCGCUGGAA--UUGCUUCCAGCGA-CAAAUUGGAAAAUUCCCACAAACAAACACAAAUGGAUGGCAAAACGCAUGCGCUGCAUUUGCUUGCCGUUGUCC .(((.((((((((((--....)))))))).-))....(((....)))..............(((((..((((((..(((.....))).))))))..)))))))). ( -29.60, z-score = -1.44, R) >consensus CGUCAUGCGUUGGAAUUUCCCGUCCAAAGAGUCGCUUGGAAAAUUCCCACACACCAAC_GCGUUG__AAGUAAAACGCAUGCGCUGAACUUUCGCCC________ ((.(((((((((........)................(((....)))..........................))))))))))...................... (-11.32 = -10.91 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:44 2011