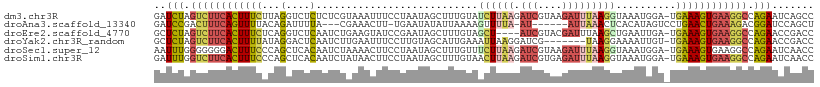
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,715,792 – 13,715,901 |
| Length | 109 |
| Max. P | 0.999666 |

| Location | 13,715,792 – 13,715,901 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.65 |
| Shannon entropy | 0.61423 |
| G+C content | 0.37542 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -13.08 |
| Energy contribution | -14.77 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
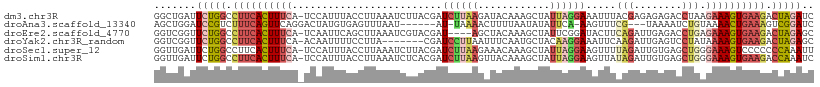
>dm3.chr3R 13715792 109 + 27905053 GGCUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUUACGAUCUUAAGAUACAAAGCUAUUAGGAAAUUUACGAGAGAGACCUAAGAAAGUGAAGACUAGAUC .......(((((.((((((((((.-..............((((((......)))))).........(((((...(((.(....))))))))))))))))))).))))).. ( -27.50, z-score = -2.71, R) >droAna3.scaffold_13340 11700657 99 - 23697760 AGCUGGAUCCGUCUUUCAGUUCAGGACUAUGUGAGUUUAAU------AU-UAAAACUUUUAAUAUAUUCA-AAGUUUCG---UAAAAUCUGUAAAACUGAAAGUCGGAUC .....((((((.(((((((((((((..((((.((.(((.((------((-(((.....)))))))....)-)).)).))---))...))))...))))))))).)))))) ( -27.30, z-score = -4.41, R) >droEre2.scaffold_4770 9837075 105 - 17746568 GGUCGGUUCUGGCCUUCACUUUCA-UCAAUUCAGCUUAAAUCGUACGAU----AGCUACAAAGCUAUUCGGAUACUUCAGAUUGAGACCUGAGAAAGUGAAGACUAGAGC .....(((((((.((((((((((.-(((......(((((.((...((((----((((....))))).)))((....)).)))))))...))))))))))))).))))))) ( -35.20, z-score = -3.66, R) >droYak2.chr3R_random 1876194 102 + 3277457 GGUCGGUUCUGGCCUUCACUUUCA-ACAAUUUUCCUUA-------CGAUCCUUAAUUUCAAUGCUACAAGGAAAUUCAAGAUUGAGUCCUAUAAAAGUGAAGACUAGAGC .....(((((((.(((((((((..-......((((((.-------......................))))))(((((....)))))......))))))))).))))))) ( -23.30, z-score = -1.45, R) >droSec1.super_12 1684430 109 - 2123299 GGUUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUUACGAUCUUAAGAAACAAAGCUAUUAGGAAGUUUUAGAUUGUGAGCUGGGAAAGUCCCCCCCAAAUU ((....(((..((..((((..((.-.........((((((((....))).)))))....((((((.......)))))).))..))))))..)))....)).......... ( -20.30, z-score = 0.35, R) >droSim1.chr3R 19746765 109 - 27517382 GGUUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUCACGAUCUUAAGUUACAAAGCUAUUAGGAAGUUAUAGAUUGUGAGCUGGGAAAGUGAAGACCAAAUC ....((((.(((.((((((((((.-.(((............(((((((((((((.((.(.((....)).).)).))).)))))))))).))))))))))))).))))))) ( -30.52, z-score = -2.74, R) >consensus GGUUGAUUCUGGCCUUCACUUUCA_UCCAUUUACCUUAAAUCUUACGAUCUUAAGAUACAAAGCUAUUAGGAAAUUUAAGAUUGAGACCUGAGAAAGUGAAGACCAGAUC .......(((((.((((((((((.........................((((((............)))))).....(((........))).)))))))))).))))).. (-13.08 = -14.77 + 1.69)
| Location | 13,715,792 – 13,715,901 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.65 |
| Shannon entropy | 0.61423 |
| G+C content | 0.37542 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.48 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
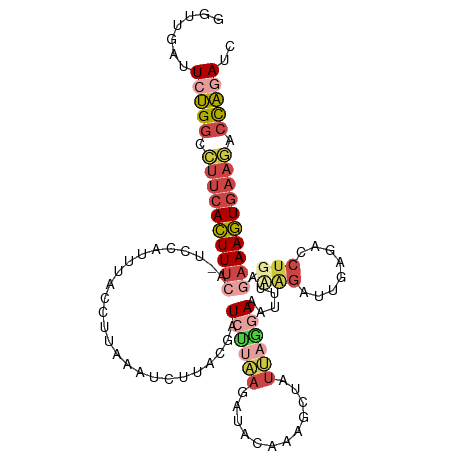
>dm3.chr3R 13715792 109 - 27905053 GAUCUAGUCUUCACUUUCUUAGGUCUCUCUCGUAAAUUUCCUAAUAGCUUUGUAUCUUAAGAUCGUAAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAGCC (((((.(((((((((((((((((................)))))...((...(((((((.((((....)))))))))))...)).-.)))))))))))).)).))).... ( -34.19, z-score = -4.16, R) >droAna3.scaffold_13340 11700657 99 + 23697760 GAUCCGACUUUCAGUUUUACAGAUUUUA---CGAAACUU-UGAAUAUAUUAAAAGUUUUA-AU------AUUAAACUCACAUAGUCCUGAACUGAAAGACGGAUCCAGCU ((((((.(((((((((...(((......---.(((((((-(..........)))))))).-..------.....(((.....))).)))))))))))).))))))..... ( -25.70, z-score = -4.89, R) >droEre2.scaffold_4770 9837075 105 + 17746568 GCUCUAGUCUUCACUUUCUCAGGUCUCAAUCUGAAGUAUCCGAAUAGCUUUGUAGCU----AUCGUACGAUUUAAGCUGAAUUGA-UGAAAGUGAAGGCCAGAACCGACC ..(((.((((((((((((((((((....))))))......(((.(((((....))))----))))..((((((.....)))))).-.)))))))))))).)))....... ( -34.10, z-score = -3.61, R) >droYak2.chr3R_random 1876194 102 - 3277457 GCUCUAGUCUUCACUUUUAUAGGACUCAAUCUUGAAUUUCCUUGUAGCAUUGAAAUUAAGGAUCG-------UAAGGAAAAUUGU-UGAAAGUGAAGGCCAGAACCGACC ..(((.((((((((((((((((((.....)))))..((((((((..(..((.......))..)..-------)))))))).....-))))))))))))).)))....... ( -27.60, z-score = -2.34, R) >droSec1.super_12 1684430 109 + 2123299 AAUUUGGGGGGGACUUUCCCAGCUCACAAUCUAAAACUUCCUAAUAGCUUUGUUUCUUAAGAUCGUAAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAACC ..(((((..(((.....)))...((((..((.......(((.....((((((..(((((......)))))..))))))....)))-.))..))))...)))))....... ( -23.40, z-score = 0.24, R) >droSim1.chr3R 19746765 109 + 27517382 GAUUUGGUCUUCACUUUCCCAGCUCACAAUCUAUAACUUCCUAAUAGCUUUGUAACUUAAGAUCGUGAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAACC (((((((((((((((((((((....((((.((((.........))))..)))).((((.(((((....))))).))))...))).-.))))))))))))))).))).... ( -38.20, z-score = -5.61, R) >consensus GAUCUAGUCUUCACUUUCACAGGUCUCAAUCUAAAACUUCCUAAUAGCUUUGUAACUUAAGAUCGUAAGAUUUAAGGUAAAUGGA_UGAAAGUGAAGGCCAGAACCAACC ..(((.((((((((((((...(....)...........................((((((.(((....)))))))))..........)))))))))))).)))....... (-13.48 = -14.45 + 0.97)
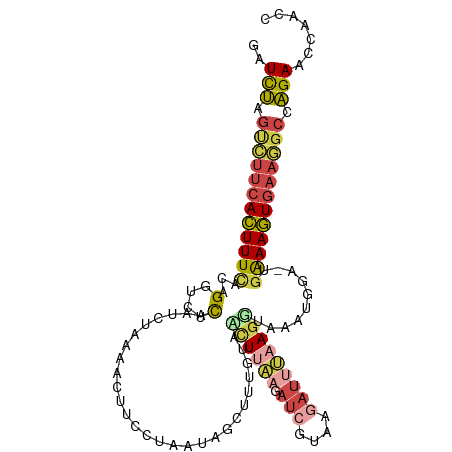
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:42 2011