| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,710,160 – 13,710,255 |
| Length | 95 |
| Max. P | 0.953458 |

| Location | 13,710,160 – 13,710,255 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.75 |
| Shannon entropy | 0.67797 |
| G+C content | 0.64677 |
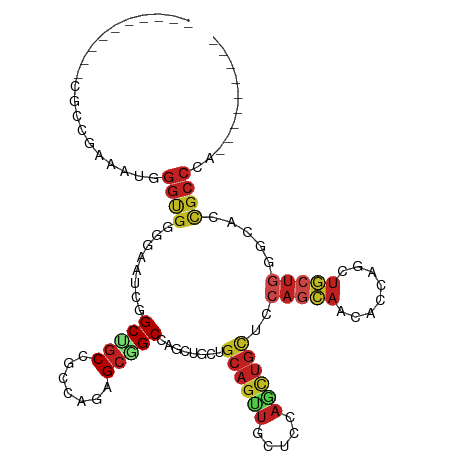
| Mean single sequence MFE | -46.14 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.81 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
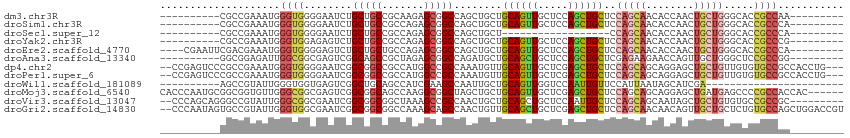
>dm3.chr3R 13710160 95 + 27905053 ----------CGCCGAAAUGGGUGGGGAAUCUGCUGCCGCAAGAGCGGCCAGCUGCUGCAGUUGCUCCAGCUGCUCCAGCAACACCAACUGCUGGGCACCGCCAA--------- ----------..........(((((.......((((((((....)))).))))(((.(((((((...))))))).((((((........))))))))))))))..--------- ( -42.60, z-score = -1.00, R) >droSim1.chr3R 19739187 95 - 27517382 ----------CGCCGAAAUGGGUGGGGAAUCUGCUGCCGCCAGAGCGGCCAGCUGCUGCAGUUGCUCCAGCUGCUCCAGCAACACCAACUGCUGGGCACCGCCCA--------- ----------........(((((((.......((((((((....)))).))))(((.(((((((...))))))).((((((........))))))))))))))))--------- ( -45.60, z-score = -1.58, R) >droSec1.super_12 1678828 77 - 2123299 ----------CGCCGAAAUGGGUGGGGAAUCUGCUGCCGCCAGAGCGGCCAGCUGCU------------------CCAGCAACACCAACUGCUGGGCACCGCCCA--------- ----------........(((((((.......((((((((....)))).))))(((.------------------((((((........))))))))))))))))--------- ( -38.80, z-score = -2.53, R) >droYak2.chr3R 2302437 95 + 28832112 ----------CGCCGAAAUGGGUGGAGAGUCUGCUGCCGCCAGAGCGGCCAGCUGCUGCAGUUGCUCCAGCUGCUCCAGCAACACCAACUGCUGGGCACCGCCCG--------- ----------.........((((((.......((((((((....)))).))))(((.(((((((...))))))).((((((........))))))))))))))).--------- ( -45.20, z-score = -1.72, R) >droEre2.scaffold_4770 9831524 101 - 17746568 ----CGAAUUCGACGAAAUGGGUGGGGAGUCUGCUGCUGCCAGAGCGGCCAGCUGCUGCAGUUGCUCCAGCUGCUCCAGCAACACCAACUGCUGGGCACCGCCCA--------- ----..............((((((((((((((((.((((((.....)).))))....))))..)))))...(((.((((((........))))))))))))))))--------- ( -46.70, z-score = -1.73, R) >droAna3.scaffold_13340 11695536 95 - 23697760 ----------GGCGGAGAUUGGCGGCGAGUCGGCAGCCGCUAGAGCGGCCAGAUGCUGCAGCUGCUCCAGCUGCUCGAGAAGAACCAGUUGCUGGGCUCCGCCGG--------- ----------(((((((.(..(((((...(((((((((((....)))))....))))(((((((...))))))).......))....)))))..).)))))))..--------- ( -50.60, z-score = -1.99, R) >dp4.chr2 28709007 109 + 30794189 --CCGAGUCCCGCCGAAAUGGGUGGGGAAUCGGCGGCCGCCAUGGCCGCCAAAUGUUGCAGUUGCUCGAGCUGCUCCAGCAGCAGGAGCUGCUGUUGUGUGCCGCCACCUG--- --.(((((((((((......)))))).....(((((((.....))))))).............))))).((.((.((((((((((...))))))))).).)).))......--- ( -52.50, z-score = -2.05, R) >droPer1.super_6 4040956 109 + 6141320 --CCGAGUCCCGCCGAAAUGGGUGGGGAAUCGGCGGCCGCCAUGGCCGCCAAAUGUUGCAGUUGCUCGAGCUGCUCCAGCAGCAGGAGCUGCUGUUGUGUGCCGCCACCUG--- --.(((((((((((......)))))).....(((((((.....))))))).............))))).((.((.((((((((((...))))))))).).)).))......--- ( -52.50, z-score = -2.05, R) >droWil1.scaffold_181089 7405744 81 - 12369635 ----------AGCCGUAUUGGGUGGUGAGUCGGCUGCAGCCAUCGAAGCCAAUUGCUGCAGUUGGUCCAAUUGUUCCAUUAAUAGCAUCGA----------------------- ----------.((..((.((((...((..(((((((((((.((.(....).)).)))))))))))..)).....)))).))...)).....----------------------- ( -26.10, z-score = -1.18, R) >droMoj3.scaffold_6540 31259789 108 - 34148556 CACCCAAUGCGGCGGUGUUGGGCGGCGAGUCGGCGGCAGCCAAGGCGGCUAGCUGCUGCAGUUGCUCGAGCUGCUCCAGCAGCAGGAGCUGAUGAGCCCCGCCACCAC------ ..........((.(((((((((((((..(.(((((((((((.....)))).)))))))).)))))))))(((((....))))).((.(((....))))))))).))..------ ( -52.20, z-score = -0.40, R) >droVir3.scaffold_13047 12220805 103 - 19223366 --CCCAGCAGGGCCGUAUUGGGCGGCGAAUCGGCGGCGGCUAAAGCCGCCAACUGCUGCAGCUGCUCCAAUUGCUCCAGCAGCAAUAGCUGCUGUGUGCCGCCGC--------- --(((....)))........(((((((...((((((((((....))))))..((((((.(((..........))).))))))........))))..)))))))..--------- ( -51.10, z-score = -1.43, R) >droGri2.scaffold_14830 3390139 112 - 6267026 --CCCAAUAGUGCCGUAUUGGGUGGCGAAUCGGCGGCGGCCAAAGCAGCCAACUGUUGCAGCUGCUCGAGCUGCUCCAGCAACAACAGUUGCUGCUCUGUGCCAGCUGGACCGU --(((((((......)))))))..(((..(((((...(((((.((((((.((((((((..((((...(((...)))))))..)))))))))))))).)).))).)))))..))) ( -49.80, z-score = -1.73, R) >consensus __________CGCCGAAAUGGGUGGGGAAUCGGCUGCCGCCAGAGCGGCCAGCUGCUGCAGUUGCUCCAGCUGCUCCAGCAACACCAGCUGCUGGGCACCGCCCA_________ ....................((((........(((((.......)))))........((((((.....))))))..(((((........))))).....))))........... (-19.47 = -19.81 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:41 2011