
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,698,963 – 13,699,058 |
| Length | 95 |
| Max. P | 0.554128 |

| Location | 13,698,963 – 13,699,058 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 52.83 |
| Shannon entropy | 1.03004 |
| G+C content | 0.38049 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -6.40 |
| Energy contribution | -5.92 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.01 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554128 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
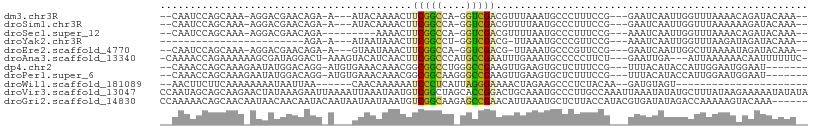
>dm3.chr3R 13698963 95 - 27905053 --CAAUCCAGCAAA-AGGACGAACAGA-A---AUACAAAACUUCGGCCA-GGUCGACGUUUAAAUGCCCUUUCCG---GAAUCAAUUGGUUUAAAACAGAUACAAA-- --...(((.(..((-(((.((...(..-.---.)...((((.(((((..-.))))).))))...)).)))))).)---)).....(((........))).......-- ( -11.40, z-score = 1.38, R) >droSim1.chr3R 19726303 95 + 27517382 --CAAUCCAGCAAA-AGGACGAACAGA-A---AUACAAAACUUCGGCCA-GGUCGACGUUUUAAUGCCCUUUCCG---GAAUCAAUUGGUUUAAAAAAGAUACAAA-- --...(((......-.)))........-.---....(((((.(((((..-.))))).)))))......((((...---(((((....)))))...)))).......-- ( -13.30, z-score = 0.78, R) >droSec1.super_12 1667640 90 + 2123299 --CAAUCCAGCAAA-AGGACGAACAGA---------AAAACUUCGGCCA-GGUCGACGUUUUAAUGCCCUUUCCG---AAAUCAAUUGGUUUAAAACAGAUACAAA-- --..(((.......-.(((....((..---------(((((.(((((..-.))))).)))))..)).....))).---(((((....)))))......))).....-- ( -13.40, z-score = 0.43, R) >droYak2.chr3R 2283107 73 - 28832112 ------------------------AGA-A---AUAAUAAACUUCGGCCU-GGUCGACG-UUAAAUGCCCGUUCCG---AAAUCAAUUGGUUUAAGAUAGAUACAAA-- ------------------------...-.---....((((((((((..(-((.((...-.....)).)))..)))---)........)))))).............-- ( -8.70, z-score = 1.13, R) >droEre2.scaffold_4770 9820119 94 + 17746568 --CAAUCCAGCAAA-AGGACGAACAGA-A---GUAAUAAACUUCGGCCA-GGUCGACG-UUAAAUGCCCGUUCCG---GAAUCAAUUGGCUUAAAAUAGAUACAAA-- --....((((((..-..((((....((-(---((.....)))))(((..-.)))..))-))...)))(((...))---).......))).................-- ( -14.30, z-score = 0.67, R) >droAna3.scaffold_13340 11683494 99 + 23697760 -CAAAACCAGAAAAAAGCGAUAGGACU-AAAGUACAUCAACUUCGGCCCAUGCCGAAUUUGAAAUGCCCCCUUCU---GAAUUGA---AUUAAAAAACAAUUUUUUC- -......(((((....(((...(.((.-...)).).((((.((((((....)))))).))))..)))....))))---)......---...................- ( -17.70, z-score = -2.09, R) >dp4.chr2 28696442 95 - 30794189 --CAAACCAGCAAAGAAUAUGGACAGG-AUGUGAAACAAACGGCGGCCUGGGCCGAAGUUGAAGUGCUCUUUCCG---UUUACAUACCAUUGGAAUGGAAU------- --....((((........(((((.(((-(.(((.....(((..((((....))))..)))....)))))))))))---)..........))))........------- ( -21.87, z-score = -0.04, R) >droPer1.super_6 4028521 95 - 6141320 --CAAACCAGCAAAGAAUAUGGACAGG-AUGUGAAACAAACGGCGGCAAGGGCCGAAGUUGAAGUGCUCUUUCCG---UUUACAUACCAUUGGAAUGGAAU------- --....((((........(((((.(((-(.(((.....(((..((((....))))..)))....)))))))))))---)..........))))........------- ( -20.37, z-score = -0.01, R) >droWil1.scaffold_181089 7393453 76 + 12369635 --AACUUCUUCAAAAAAAAUAAUUAA------CAACAAAAAAUCCCUCAUUAGGGAAAACUAGAAGCCCUCUACAA--GAUGUAGU---------------------- --.((.((((................------..........(((((....)))))....((((.....)))).))--)).))...---------------------- ( -9.20, z-score = -1.36, R) >droVir3.scaffold_13047 12207974 108 + 19223366 CCAAUAGCAGCAAGAACUAUAAAGAAUUAAAAUUAAAUAAUGUCGGCUAGCACCGGACUGCAAAUGCCCUUGCCAAAUUAAAUAUAUGCUUUAUAAGAAAAAUAUAUA .........(((((.........((((((........)))).))(((..(((......)))....))))))))........((((((..(((....)))..)))))). ( -13.40, z-score = 0.66, R) >droGri2.scaffold_14830 3378869 102 + 6267026 CCAAAAACAGCAACAAUAACAACAAUACAAUAAUAAUAAAUGUCGGCAAGAGCCGAACAUUAAAUGCUCUUACCAUACGUGAUAUAGACCAAAAAGUACAAA------ .........................(((..........(((((((((....)))).)))))...((.(((((.((....)).)).))).))....)))....------ ( -12.00, z-score = -1.70, R) >consensus __CAAACCAGCAAA_AGGACGAACAGA_A___AUAAAAAACUUCGGCCA_GGCCGAAGUUUAAAUGCCCUUUCCG___GAAUCAAUUGGUUUAAAAGAGAUACAAA__ ..........................................(((((....))))).................................................... ( -6.40 = -5.92 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:37 2011