| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,833,692 – 7,833,785 |
| Length | 93 |
| Max. P | 0.993947 |

| Location | 7,833,692 – 7,833,785 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.69702 |
| G+C content | 0.60571 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.67 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993947 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
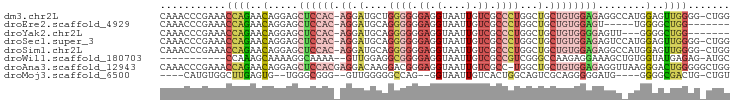
>dm3.chr2L 7833692 93 + 23011544 CAAACCCGAAACCAGAACAGGAGCUCCAC-AGGAUGCUGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGAGAGGCCAUGGAGUUGGGG-CUGG ((..(((((..(((.....((..((((((-((.(.((..(((.((.(....).)).)))..))).))))))))...)).)))..))))).-.)). ( -46.60, z-score = -4.01, R) >droEre2.scaffold_4929 16755132 82 + 26641161 CAAACCCGAAACCAGAACAGGAGCUCCAC-AGGAUGCAGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGAGU-----UGGGGCUGG------- ((..(((....((......))((((((((-((.(.((.((((.((.(....).)).)))).))).)))))))))-----))))..)).------- ( -38.40, z-score = -3.05, R) >droYak2.chr2L 17260641 84 - 22324452 CAAACCCGAAACCAGAACAGGAGCUCCAC-AGGAUGCAGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGGGAGUU---GGGGCUGG------- ((..(((((..((......))..((((((-((.(.((.((((.((.(....).)).)))).))).))))))))..))---)))..)).------- ( -36.60, z-score = -2.11, R) >droSec1.super_3 3347887 93 + 7220098 CAAACCCGAAACCAGAACAGGAGCUCCAC-AGGAUGCAGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGAGAGUCCAUGGAGUUGGGG-CUGG ((..(((((..(((.....(((.((((((-((.(.((.((((.((.(....).)).)))).))).))))))))..))).)))..))))).-.)). ( -44.30, z-score = -3.52, R) >droSim1.chr2L 7629790 93 + 22036055 CAAACCCGAAACCAGAACAGGAGCUCCAC-AGGAUGCAGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGAGAGGCCAUGGAGUUGGGG-CUGG ((..(((((..(((.....((..((((((-((.(.((.((((.((.(....).)).)))).))).))))))))...)).)))..))))).-.)). ( -43.40, z-score = -3.19, R) >droWil1.scaffold_180703 442908 81 - 3946847 -----------CCAAAGCAAAAGGCAAAA--GUUGGAGGCGGGGAGGUAAUUGUCGCCGUCGGGCCAAGAGGAAAGCUGUGGUAUGAGAG-AUGC -----------(((.(((....(((....--)))((.(((((.((.(....).)).)))))...)).........))).)))........-.... ( -20.70, z-score = -1.18, R) >droAna3.scaffold_12943 3566033 94 - 5039921 CAAACCCGAAACCAGAACAGGAGCUCCACGAGGACAAGGACGGGAGGUAAUUGUCGCC-UGGCUGCUGUGGAGAGGUUAAGGGACUGGGGGCUGG ((..(((.(..((..(((.....(((((((.((...(((.(((.((....)).)))))-)..))..)))))))..)))...))..).)))..)). ( -31.00, z-score = -0.60, R) >droMoj3.scaffold_6500 29605501 80 - 32352404 ----CAUGUGGCUUGAGUG--UGGGCGGG--GUUGGGGGCCAG--GGUAAUUGUCACUGGCAGUCGCAGGGGGAUG----GGGGCGACUG-CUGU ----...(((((...(.(.--(((.(...--......).))).--).)....))))).(((((((((.........----...)))))))-)).. ( -26.60, z-score = -1.08, R) >consensus CAAACCCGAAACCAGAACAGGAGCUCCAC_AGGAUGCAGGGGGGAGGUAAUUGUCGCCCUGGCUGCUGUGGAGAGGC_AUGGGGCUGGGG_CUGG ...........((((..(.....((((((..((.....((((.((.(....).)).)))).....))))))))........)..))))....... (-13.61 = -14.67 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:26 2011