| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,669,967 – 13,670,063 |
| Length | 96 |
| Max. P | 0.748393 |

| Location | 13,669,967 – 13,670,063 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.73760 |
| G+C content | 0.53342 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.29 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
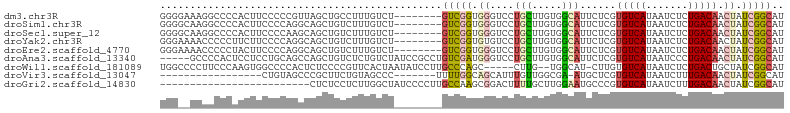
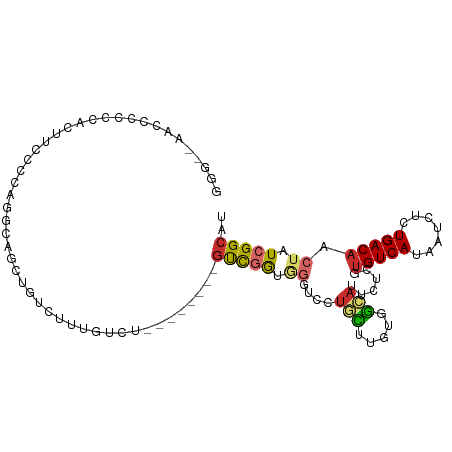
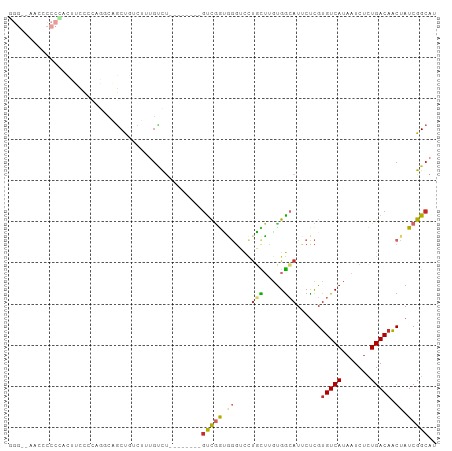
>dm3.chr3R 13669967 96 + 27905053 GGGGAAAGGCCCCACUUCCCCCGUUAGCUGCCUUUGUCU--------GUCGGUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU ((((.(((......))).))))................(--------(((((((((((..(.(((((((((....))))))))).)...)))..))))))))). ( -31.70, z-score = -1.44, R) >droSim1.chr3R 19697245 96 - 27517382 GGGGCAAGGCCCCACUUCCCCAGGCAGCUGUCUUUGUCU--------GUCGGUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU ((((.(((......)))))))((((((......))))))--------(((((((((((..(.(((((((((....))))))))).)...)))..)))))))).. ( -34.90, z-score = -1.83, R) >droSec1.super_12 1638889 96 - 2123299 GGGGCAAGGCCCCACUUCCCCAAGCAGCUGUCUUUGUCU--------GUCGGUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU ((((.(((......))))))).................(--------(((((((((((..(.(((((((((....))))))))).)...)))..))))))))). ( -31.40, z-score = -1.19, R) >droYak2.chr3R 2252933 96 + 28832112 GGGAAAACCCCCUUCUUCCCCAGGCAGCUGUCUUUGUCU--------GUCGGUGUGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU (((((..........))))).((((((......))))))--------(((((((((((..(.(((((((((....))))))))).)...))))..))))))).. ( -28.40, z-score = -2.34, R) >droEre2.scaffold_4770 9791160 96 - 17746568 GGGAAAACCCCCUACUUCCCCAGGCAGCUGUCUUUGUCU--------GUCGGUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU (((((..........))))).((((((......))))))--------(((((((((((..(.(((((((((....))))))))).)...)))..)))))))).. ( -29.40, z-score = -1.64, R) >droAna3.scaffold_13340 11654897 99 - 23697760 -----GCCCCACUCCUCCUGCAGCCAGCUGUCUCUGUCUAUCCGCCUGUCGAUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCCCUGACAACUAUCGGCAU -----((............((((....))))............)).((((((((((((..(.(((((((((....))))))))).)...)))..))))))))). ( -24.65, z-score = -0.95, R) >droWil1.scaffold_181089 1706642 96 - 12369635 UGGCCCCUUCCCAAGUGGCCCCACUCUCCCCGUUCACUAAUAUCCUUGCCCAGC-----CUUG--UGGCAU-CUUGUGUCAUAAUCUCUGACUGCUAUCGGCAU .((((.((.....)).))))..........................((((..((-----(...--.)))..-...((((((.......)))).))....)))). ( -20.10, z-score = -0.74, R) >droVir3.scaffold_13047 12173615 79 - 19223366 -----------------CUGUAGCCCGCUUCUGUAGCCC-------UUUUGGCAGCAUUUGUUGGCGA-AUGCUCGUGUCAUAAUCUUUGACAACUAUCGGCAU -----------------.....(((..........(((.-------....)))((((((((....)))-)))))..(((((.......)))))......))).. ( -21.80, z-score = -1.17, R) >droGri2.scaffold_14830 3345703 79 - 6267026 -------------------------CUCUCCUCUUGGCUAUCCCCUUGCCAAGCGGACUUUUGCUUGGAAUGCCCGUGUCAUAAUCUUUGACAACUAUCGGCAU -------------------------...(((.((((((.........)))))).)))............(((((..(((((.......)))))......))))) ( -23.00, z-score = -3.18, R) >consensus GGG__AACCCCCCACUUCCCCAGGCAGCUGUCUUUGUCU________GUCGGUGGGUCCUGCUUGUGGCAUUCUCGUGUCAUAAUCUCUGACAACUAUCGGCAU ...............................................(((((.((....(((.....)))......(((((.......))))).)).))))).. ( -8.10 = -8.29 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:35 2011