
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,829,754 – 7,829,856 |
| Length | 102 |
| Max. P | 0.700652 |

| Location | 7,829,754 – 7,829,856 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 64.52 |
| Shannon entropy | 0.71236 |
| G+C content | 0.37383 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -6.79 |
| Energy contribution | -8.10 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7829754 102 - 23011544 UGGUGAAUGACAUG----ACAGAAUAUCUGCGAAACAGAAAGUACUGAGAAAACCGAAAGUGAU-----------------CCGCAUGU-GCGGAUGAUAAAACCUUGUCAAAGGCCAAGGUUA .(((........((----.(((.....)))))...(((......))).....)))....((.((-----------------((((....-)))))).))..(((((((........))))))). ( -25.40, z-score = -2.41, R) >droSim1.chr2L 7625851 117 - 22036055 UGGUGAAUGACAUG----ACAGAAUAUCUGCGAAACAGAAAGUACUGAGAACACCGAAAGUGAUUUCUAAAAUAAAU--GACAGAAUGU-GCGGAUGAUAAAACCUUGUCAAAGGGCAAGGUUA ..............----......((((((((...(((......)))((((((.(....)))..)))).........--.........)-)))))))....(((((((((....))))))))). ( -25.40, z-score = -2.18, R) >droSec1.super_3 3343927 117 - 7220098 UGGUGAAUGACAUG----ACAAAAUAUCUGCGAAACAGAAAGUACUGAGAAAACCGAAAGUGAUUUCUAAAAUAAGU--GACAGAAUGU-GCGGAUGAUAAAACCUUGUCAAAGGGCAAGGUUA ..............----......(((((((...(((.....((((.(((((.((....).).)))))......)))--)......)))-)))))))....(((((((((....))))))))). ( -27.80, z-score = -3.21, R) >droYak2.chr2L 17256669 119 + 22324452 UGGUGUUUAUCAUA----ACAGAAUAUCUGCGAAACAGAAAGUAUUGAGGAAACCGAAAGUGAUUUCUAAAAUAAAUGCGACGACAUGU-GCGGUUGAAAAAACCUUGUCAAAGGGCAAGGUUU ..((((((((....----........((((.....))))........(((((.((....).).)))))...))))))))..((((....-...))))...((((((((((....)))))))))) ( -25.90, z-score = -1.31, R) >droEre2.scaffold_4929 16751123 121 - 26641161 UGGUGAAUAACAUGAAUAACAGAAUAUCUGCGAAACAGAAAGUAUUGAGGAAACCGAAAGUGCUUUCCAAAAUAAAU--GACACCAUGU-GUGGAUGAUAAAACCUUGUAAAAGGGCAAGGUUA (((((..............(((.....))).......(((((((((..(....)....)))))))))..........--..)))))...-...........((((((((......)))))))). ( -28.10, z-score = -2.86, R) >droAna3.scaffold_12943 3562183 119 + 5039921 UGGAGCAUGACUGG----ACAGAA-AUGUUUAGAAAACGAACUCUGGCGAAAACUGAAAGUGAAUCCGAAAAUAACUUGGAACUAGUAUGGUGGGAGGUGGAACCUUGCGAGAGGCUAAGGCCA .((((..((.((((----(((...-.)))))))....))..))))(((................(((((.......)))))..((((....((.((((.....)))).))....))))..))). ( -29.20, z-score = -1.69, R) >droMoj3.scaffold_6540 31198549 99 + 34148556 --------------------GGUGUACAUAUAUAAAAAUGAAUAUGCAUGAAUAUAAAAAUUAG---AAAAUUAAGC--UUUUUGGUUGGCCGUAUACAUAAAUUGAACAAAAAGCCCAGCUUA --------------------.((((((.(((((....(((......)))..)))))..((((((---(((.......--)))))))))....))))))..............((((...)))). ( -12.20, z-score = 0.87, R) >consensus UGGUGAAUGACAUG____ACAGAAUAUCUGCGAAACAGAAAGUAUUGAGAAAACCGAAAGUGAUUUCUAAAAUAAAU__GACAGAAUGU_GCGGAUGAUAAAACCUUGUCAAAGGGCAAGGUUA ........................(((((((....(((......))).......(....)..............................)))))))....(((((((........))))))). ( -6.79 = -8.10 + 1.31)

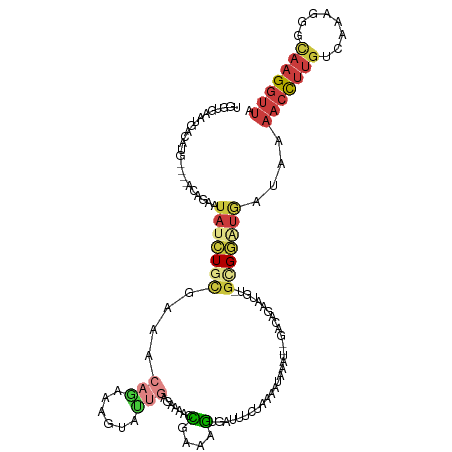

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:25 2011