| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,630,964 – 13,631,055 |
| Length | 91 |
| Max. P | 0.575014 |

| Location | 13,630,964 – 13,631,055 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 62.32 |
| Shannon entropy | 0.70303 |
| G+C content | 0.44475 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -5.43 |
| Energy contribution | -6.93 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
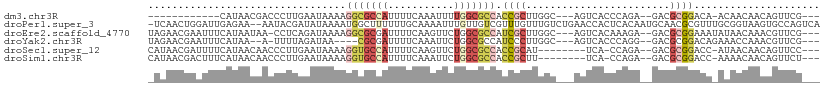
>dm3.chr3R 13630964 91 - 27905053 ------------CAUAACGACCCUUGAAUAAAAGGCGCCAUUUUCAAAUUUUGGCGCCACCGCUUGGC---AGUCACCCAGA--GACGCGGACA-ACAACAACAGUUCG--- ------------.............((((....(((((((...........))))))).((((((((.---......)))).--...))))...-.........)))).--- ( -24.00, z-score = -2.33, R) >droPer1.super_3 4041754 109 + 7375914 -UCAACUGGAUUGAGAA--AAUACGAUAUAAAAUGGCUUUUUUGCAAAAUUUGUUGUCGUUUGUUUGUCUGAACCACUCACAAUGCAACGCGUUUGCGGUAAGUGCCAGUCA -...(((((...((.((--(..((((((..((((.((......))...))))..))))))...))).)).....((((.((...((((.....)))).)).))))))))).. ( -25.90, z-score = -0.54, R) >droEre2.scaffold_4770 9747950 103 + 17746568 UAGAACGAAUUUCAUAAUAA-CCUCAGAUAAAAGGCGCGAUUUUCAAGUUCUGGCGCCAUCGCUUGGC---AGUCACAAAGA--GACGCGGAAAUAUAACAAACGUUCG--- ..(((((.............-(((........)))((((..((((...((.((((((((.....))))---.)))).)).))--)))))).............))))).--- ( -24.30, z-score = -1.63, R) >droYak2.chr3R 2212078 97 - 28832112 UAGAACGAAUUUCAUAA--A-UUUUAGAUAA----CGCGAUUUUCAAAUUCUGGCGCCAUCCCUUGGC---AGUCACCCAGG--GACGCGGACAGAAACCAAACGUUCG--- ..(((((((..((....--.-..........----...))..)))...(((((.(((..(((((.((.---.....)).)))--)).)))..))))).......)))).--- ( -25.37, z-score = -2.40, R) >droSec1.super_12 1599909 97 + 2123299 CAUAACGAUUUUCAUAACAACCCUUGAAUAAAAGGUGCCAUUUUCAAGUUCUGGCGCCACCGCAU--------UCA-CCAGA--GACGCGGACC-AUAACAACAGUUCC--- ..................(((..(((.......(((((((...........))))))).((((.(--------((.-...))--)..))))...-....)))..)))..--- ( -19.60, z-score = -1.84, R) >droSim1.chr3R 19657828 97 + 27517382 CAUAACGACUUUCAUAACAACCCUUGAAUAAAAGGUGCCAUUUUCAAAUUCUGGCGCCACCGCUU--------UCA-CCAGA--GACGCGGACC-AAAACAACAGUUCU--- .........................((((....(((((((...........))))))).((((((--------((.-...))--)).))))...-.........)))).--- ( -22.60, z-score = -3.18, R) >consensus _AGAACGAAUUUCAUAACAACCCUUAAAUAAAAGGCGCCAUUUUCAAAUUCUGGCGCCACCGCUUGGC___AGUCACCCAGA__GACGCGGACA_AAAACAACAGUUCG___ .................................(((((((...........))))))).((((........................))))..................... ( -5.43 = -6.93 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:27 2011