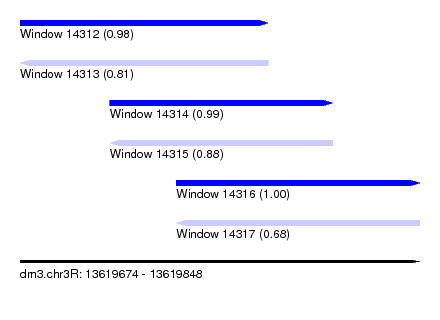
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,619,674 – 13,619,848 |
| Length | 174 |
| Max. P | 0.998603 |

| Location | 13,619,674 – 13,619,782 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.30839 |
| G+C content | 0.50955 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -22.30 |
| Energy contribution | -24.63 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
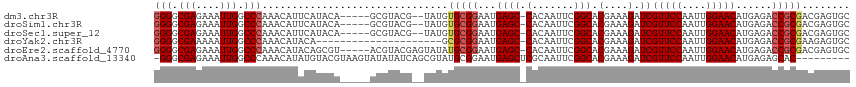
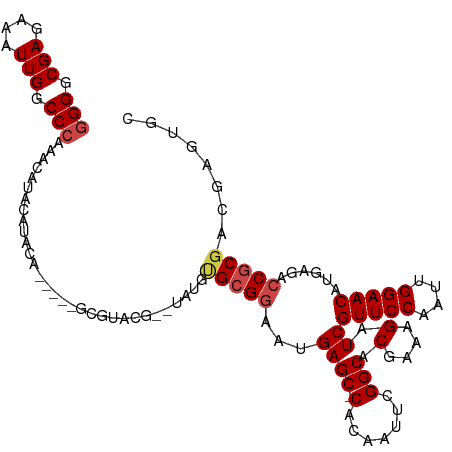
>dm3.chr3R 13619674 108 + 27905053 GGGGCGAGAAAUUGGCCCAAACAUUCAUACA-----GCGUACG--UAUGUGCGGAAUGAGC-CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGC (((.(((....))).)))...(((((((((.-----......)--)))((((((...((((-(.......))).(....).))(((((....)))))......)))).))))))). ( -36.90, z-score = -2.72, R) >droSim1.chr3R 19646626 108 - 27517382 GGGGCGAGAAAUUGGCCCAAACAUUCAUACA-----GCGUACG--UAUGUGCGGAAUGAGC-CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGC (((.(((....))).)))...(((((((((.-----......)--)))((((((...((((-(.......))).(....).))(((((....)))))......)))).))))))). ( -36.90, z-score = -2.72, R) >droSec1.super_12 1588793 108 - 2123299 GGGGCGAGAAAUUGGCCCAAACAUUCAUACA-----GCGUACG--UAUGUGCGGAAUGAGC-CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGC (((.(((....))).)))...(((((((((.-----......)--)))((((((...((((-(.......))).(....).))(((((....)))))......)))).))))))). ( -36.90, z-score = -2.72, R) >droYak2.chr3R 2199117 94 + 28832112 GGGGCGAAAAAUUGGCCCAAACAUACA---------------------GCGCGGAAUGAGC-CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGAAGAGUGC (((.(((....))).)))...(((.(.---------------------.(((((...((((-(.......))).(....).))(((((....)))))......)))))..).))). ( -31.70, z-score = -3.12, R) >droEre2.scaffold_4770 9735133 110 - 17746568 GGGGCGAGAAAUUGGCCCAAACAUACAGCGU-----ACGUACGAGUAUAUGCGGAAUGAGC-CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGC (((.(((....))).))).............-----..((((..((...(((((...((((-(.......))).(....).))(((((....)))))......)))))))..)))) ( -35.30, z-score = -2.65, R) >droAna3.scaffold_13340 12914770 106 - 23697760 -GGGCGAGAAAUUGGCCCAAACAUAUGUACGUAAGUAUAUAUCAGCGUAUGCGGAAUGAGCUCGCAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGAGCAC--------- -((((((....)).)))).....(((((((....)))))))(((.((....))...)))((((((......)).(....)...(((((....)))))....))))..--------- ( -33.50, z-score = -3.07, R) >consensus GGGGCGAGAAAUUGGCCCAAACAUACAUACA_____GCGUACG__UAUGUGCGGAAUGAGC_CACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGC (((.(((....))).)))...(((........................((((.((((.........)))).))))........(((((....))))))))................ (-22.30 = -24.63 + 2.33)
| Location | 13,619,674 – 13,619,782 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.30839 |
| G+C content | 0.50955 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13619674 108 - 27905053 GCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG-GCUCAUUCCGCACAUA--CGUACGC-----UGUAUGAAUGUUUGGGCCAAUUUCUCGCCCC ((..(((.(((((.....((((((....)))))).....))))).)))....((-(((((..(....((((--((.....-----))))))...)..))))))).......))... ( -32.50, z-score = -1.63, R) >droSim1.chr3R 19646626 108 + 27517382 GCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG-GCUCAUUCCGCACAUA--CGUACGC-----UGUAUGAAUGUUUGGGCCAAUUUCUCGCCCC ((..(((.(((((.....((((((....)))))).....))))).)))....((-(((((..(....((((--((.....-----))))))...)..))))))).......))... ( -32.50, z-score = -1.63, R) >droSec1.super_12 1588793 108 + 2123299 GCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG-GCUCAUUCCGCACAUA--CGUACGC-----UGUAUGAAUGUUUGGGCCAAUUUCUCGCCCC ((..(((.(((((.....((((((....)))))).....))))).)))....((-(((((..(....((((--((.....-----))))))...)..))))))).......))... ( -32.50, z-score = -1.63, R) >droYak2.chr3R 2199117 94 - 28832112 GCACUCUUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG-GCUCAUUCCGCGC---------------------UGUAUGUUUGGGCCAAUUUUUCGCCCC (((..(..(((((......((..(((((((.((((....)))).)).)))))..-)).....))))).---------------------.)..)))..((((.........)))). ( -29.50, z-score = -2.38, R) >droEre2.scaffold_4770 9735133 110 + 17746568 GCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG-GCUCAUUCCGCAUAUACUCGUACGU-----ACGCUGUAUGUUUGGGCCAAUUUCUCGCCCC ((..(((.(((((.....((((((....)))))).....))))).)))....((-(((((....(((((((..(((....-----))).))))))).))))))).......))... ( -34.30, z-score = -2.51, R) >droAna3.scaffold_13340 12914770 106 + 23697760 ---------GUGCUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGCGAGCUCAUUCCGCAUACGCUGAUAUAUACUUACGUACAUAUGUUUGGGCCAAUUUCUCGCCC- ---------((((......(((((((((((.((((....)))).)).))))).))))....((.((....)).))...........))))........((((.........))))- ( -24.60, z-score = -0.91, R) >consensus GCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUG_GCUCAUUCCGCACAUA__CGUACGC_____UGUAUGAAUGUUUGGGCCAAUUUCUCGCCCC .........((((......((.((((((((.((((....)))).)).))))).).)).....))))................................((((.........)))). (-22.37 = -22.40 + 0.03)

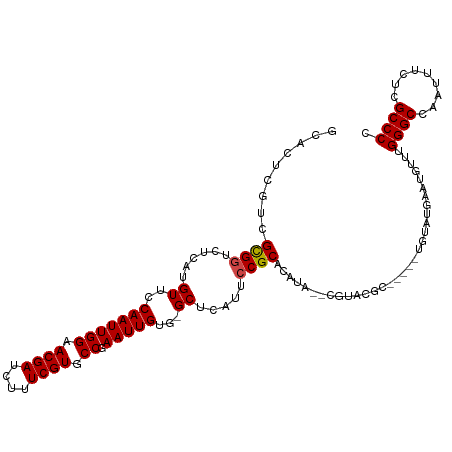
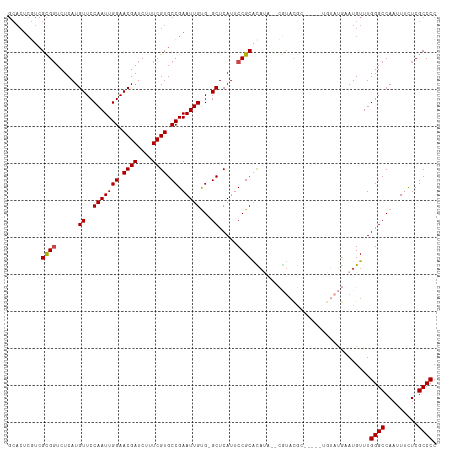
| Location | 13,619,713 – 13,619,810 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Shannon entropy | 0.22086 |
| G+C content | 0.52373 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13619713 97 + 27905053 ------AUGUGCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCACUCGU------------ ------.(((((.((((........)))).))))).......(((((....)))))(((((...((((((((((((......))))))))...)))).)))))------------ ( -36.30, z-score = -2.95, R) >droSim1.chr3R 19646665 97 - 27517382 ------AUGUGCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCACUCGU------------ ------.(((((.((((........)))).))))).......(((((....)))))(((((...((((((((((((......))))))))...)))).)))))------------ ( -36.30, z-score = -2.95, R) >droSec1.super_12 1588832 97 - 2123299 ------AUGUGCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCACUCGU------------ ------.(((((.((((........)))).))))).......(((((....)))))(((((...((((((((((((......))))))))...)))).)))))------------ ( -36.30, z-score = -2.95, R) >droYak2.chr3R 2199146 105 + 28832112 ----------GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGAAGAGUGCACCUCCAUGCAUUCGCAAUCGCAUUUGCACUCGCACUCGU ----------..((((((((((.......))).(....).))))))).........(((((...((((.(((((((......)))))))((((......))))..)))).))))) ( -36.30, z-score = -2.61, R) >droEre2.scaffold_4770 9735168 114 - 17746568 ACGAGUAUAUGCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAUC-GCACUCGCAUUCGU ((((((....((((...(((((.......))).(....).))(((((....)))))......))))(.(((((((......((((....)))).......-)))))))))))))) ( -39.42, z-score = -2.46, R) >consensus ______AUGUGCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCACUCGU____________ ..........((((((((((((.......))).(....).))))))).................((((((((((((......))))))))...))))....))............ (-30.90 = -30.86 + -0.04)

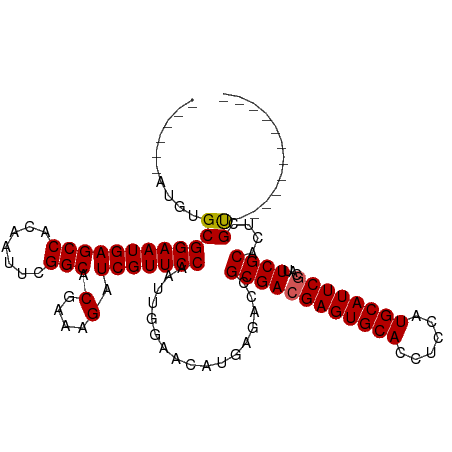
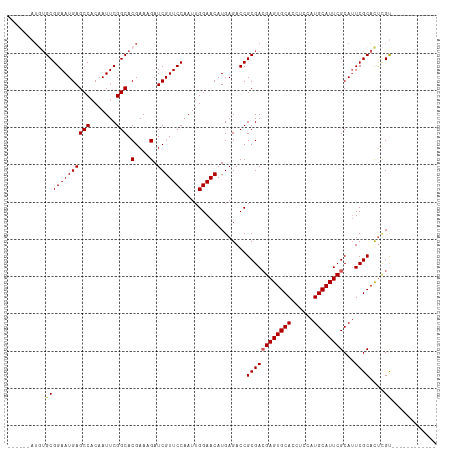
| Location | 13,619,713 – 13,619,810 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.36 |
| Shannon entropy | 0.22086 |
| G+C content | 0.52373 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13619713 97 - 27905053 ------------ACGAGUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGCACAU------ ------------(((((((((.((((....)))).....))))))))).((((......((..(((((((.((((....)))).)).)))))..)).....))))....------ ( -37.50, z-score = -2.13, R) >droSim1.chr3R 19646665 97 + 27517382 ------------ACGAGUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGCACAU------ ------------(((((((((.((((....)))).....))))))))).((((......((..(((((((.((((....)))).)).)))))..)).....))))....------ ( -37.50, z-score = -2.13, R) >droSec1.super_12 1588832 97 + 2123299 ------------ACGAGUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGCACAU------ ------------(((((((((.((((....)))).....))))))))).((((......((..(((((((.((((....)))).)).)))))..)).....))))....------ ( -37.50, z-score = -2.13, R) >droYak2.chr3R 2199146 105 - 28832112 ACGAGUGCGAGUGCAAAUGCGAUUGCGAAUGCAUGGAGGUGCACUCUUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC---------- ..(((((((.(((((..(((....)))..))))).....)))))))...((((......((..(((((((.((((....)))).)).)))))..)).....))))---------- ( -37.50, z-score = -1.35, R) >droEre2.scaffold_4770 9735168 114 + 17746568 ACGAAUGCGAGUGC-GAUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGCAUAUACUCGU ((((..((((((((-(...(..((((....)))).)...))))))))).((((......((..(((((((.((((....)))).)).)))))..)).....))))......)))) ( -41.40, z-score = -1.34, R) >consensus ____________ACGAGUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGCACAU______ ............(((((((((.((((....)))).....))))))))).((((......((..(((((((.((((....)))).)).)))))..)).....)))).......... (-32.16 = -32.96 + 0.80)


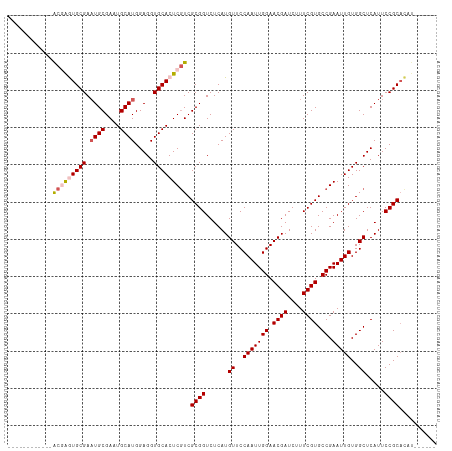
| Location | 13,619,742 – 13,619,848 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.87 |
| Shannon entropy | 0.14835 |
| G+C content | 0.43192 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13619742 106 + 27905053 AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAC------------UCGUAUUCAUACUUAUUCAUACAUAAGUCCAAAGAACUUUCA ...((..((((...(((((..(((((...((((((((((((......))))))))...)))).)------------)))).......(((((......)))))))))).))))..)). ( -29.90, z-score = -4.38, R) >droSim1.chr3R 19646694 102 - 27517382 AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAC------------UCGUAUUCAUACUUAUUCAUA----AGUCCAAAGAACUUUCA ...((..((((...(((((..(((((...((((((((((((......))))))))...))))..------------..(((....)))...))))).----..))))).))))..)). ( -29.00, z-score = -4.09, R) >droSec1.super_12 1588861 102 - 2123299 AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAC------------UCGUAUUCAUACUUAUUCAUA----AGUCCAAAGAACUUUCA ...((..((((...(((((..(((((...((((((((((((......))))))))...))))..------------..(((....)))...))))).----..))))).))))..)). ( -29.00, z-score = -4.09, R) >droYak2.chr3R 2199171 114 + 28832112 AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGAAGAGUGCACCUCCAUGCAUUCGCAAUCGCAUUUGCACUCGCACUCGUAUUCAUACUUAUUCAUA----AGUCCAAAGAACUUUCA ...((..((((...(((((..(((((...((((.(((((((......)))))))((((......))))..))))....(((....)))...))))).----..))))).))))..)). ( -32.10, z-score = -3.93, R) >droEre2.scaffold_4770 9735203 113 - 17746568 AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAUC-GCACUCGCAUUCGUAUUCAUACUUAUUCAUA----AGUCCAAAGAACUUUCA ...((..((((...(((((..(((((...((((((((((((......))))))))...))))...-((....)).................))))).----..))))).))))..)). ( -30.60, z-score = -3.44, R) >consensus AAAGAUCGUUCCAAUUGGAACAUGAGACCGCGACGAGUGCACCUCCAUGCAUUCGCAUUCGCAC____________UCGUAUUCAUACUUAUUCAUA____AGUCCAAAGAACUUUCA ((((.(((((((....))))).((.(((.((((((((((((......))))))))...))))................(((....)))..............)))))..)).)))).. (-26.82 = -27.02 + 0.20)

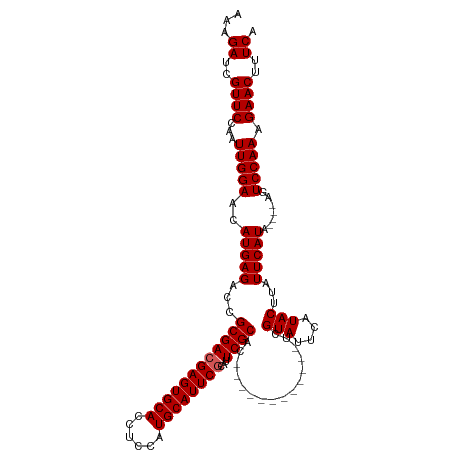
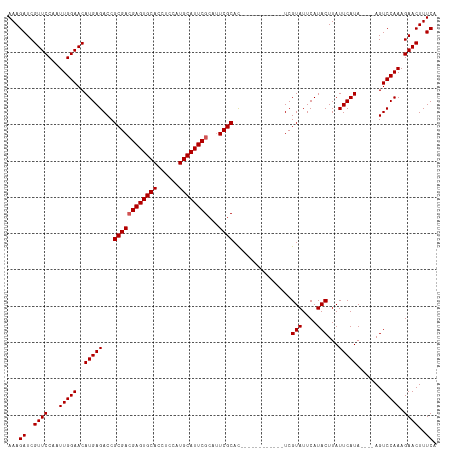
| Location | 13,619,742 – 13,619,848 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.87 |
| Shannon entropy | 0.14835 |
| G+C content | 0.43192 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13619742 106 - 27905053 UGAAAGUUCUUUGGACUUAUGUAUGAAUAAGUAUGAAUACGA------------GUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..(((((((((((((((......)))))(((((....((------------.((((...((((.(((((....))))).)))))))).))))))).)))))..)))))..))... ( -31.60, z-score = -1.98, R) >droSim1.chr3R 19646694 102 + 27517382 UGAAAGUUCUUUGGACU----UAUGAAUAAGUAUGAAUACGA------------GUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..((((((((((..----(((((....(((....)))((------------.((((...((((.(((((....))))).)))))))).))))))).)))))..)))))..))... ( -31.90, z-score = -2.39, R) >droSec1.super_12 1588861 102 + 2123299 UGAAAGUUCUUUGGACU----UAUGAAUAAGUAUGAAUACGA------------GUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..((((((((((..----(((((....(((....)))((------------.((((...((((.(((((....))))).)))))))).))))))).)))))..)))))..))... ( -31.90, z-score = -2.39, R) >droYak2.chr3R 2199171 114 - 28832112 UGAAAGUUCUUUGGACU----UAUGAAUAAGUAUGAAUACGAGUGCGAGUGCAAAUGCGAUUGCGAAUGCAUGGAGGUGCACUCUUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..((((((((((..----(((((....(((....)))((.((((((((((..(((....)))..)))))((((.....))))))))).))))))).)))))..)))))..))... ( -34.80, z-score = -1.99, R) >droEre2.scaffold_4770 9735203 113 + 17746568 UGAAAGUUCUUUGGACU----UAUGAAUAAGUAUGAAUACGAAUGCGAGUGC-GAUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..((((((((((..----(((((....((((........))))((.(((-(((((....))((.(((((....))))).)))))))).))))))).)))))..)))))..))... ( -35.20, z-score = -1.98, R) >consensus UGAAAGUUCUUUGGACU____UAUGAAUAAGUAUGAAUACGA____________GUGCGAAUGCGAAUGCAUGGAGGUGCACUCGUCGCGGUCUCAUGUUCCAAUUGGAACGAUCUUU .((..((((((((((...............(((....)))..............(((.((.(((((.(((((....)))))....))))).)).)))..)))))..)))))..))... (-24.94 = -24.70 + -0.24)

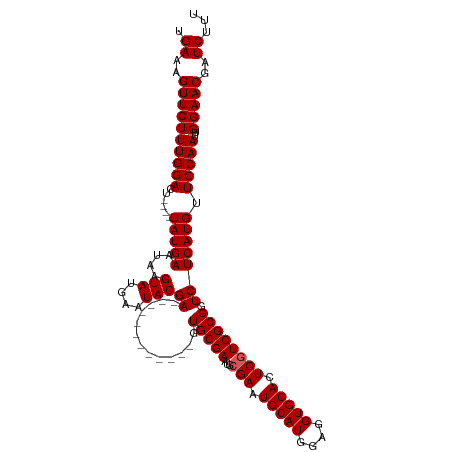
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:26 2011