
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,618,731 – 13,618,863 |
| Length | 132 |
| Max. P | 0.762614 |

| Location | 13,618,731 – 13,618,829 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Shannon entropy | 0.49111 |
| G+C content | 0.52082 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.57 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
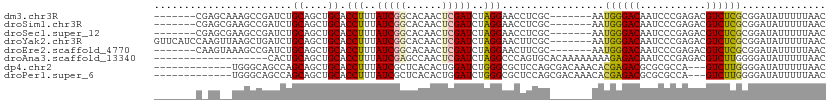
>dm3.chr3R 13618731 98 - 27905053 -------CGAGCAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC -------((((....((((((.(((....))).....))))))....))))...((((((..((((-------...(((.....)))(((....)))))))...)))))).. ( -28.40, z-score = -1.79, R) >droSim1.chr3R 19645688 98 + 27517382 -------CGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC -------((((....((((((.(((....))).....))))))....))))...((((((..((((-------...(((.....)))(((....)))))))...)))))).. ( -28.40, z-score = -1.38, R) >droSec1.super_12 1587860 98 + 2123299 -------CGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC -------((((....((((((.(((....))).....))))))....))))...((((((..((((-------...(((.....)))(((....)))))))...)))))).. ( -28.40, z-score = -1.38, R) >droYak2.chr3R 2197689 105 - 28832112 GUUCAUCCAAGUUAAGCUGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC ....(((((((((.(((((.....)))))...(((..(((((......)))))..)))))))).((-------...(((.....)))(((....)))))))))......... ( -30.80, z-score = -2.70, R) >droEre2.scaffold_4770 9734196 98 + 17746568 -------CAAGUAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC -------........((((((.(((....))).....))))))...........((((((.(((((-------...(((.....)))(((....))))))))..)))))).. ( -25.60, z-score = -1.43, R) >droAna3.scaffold_13340 12913556 93 + 23697760 -------------------CACUGCAGCUGCACCUUUAUCGAGCCAACUCGAUCUAGGCCCAGUGCACAAAAAAAAGAGACAAUCCCGAGACGUCUUGGGGAUAUUUUUAAC -------------------...((((.(((..(((..((((((....))))))..)))..)))))))........(((((.(.(((((((....))))))).).)))))... ( -29.30, z-score = -3.77, R) >dp4.chr2 25969142 96 - 30794189 -------------UGGGCAGCCAGCAGCUGCACCUUUAUCGCUCACACUGGAUCUGGGCGCUCCAGCGACAAACACGAGACGCGCGCCA---GUCUUGGGGAUAUUUUUAAC -------------.(((((((.....))))).))..((((.((((....(((.((((((((((...((.......)).)).)))).)))---))))))))))))........ ( -31.90, z-score = -1.32, R) >droPer1.super_6 1302705 96 - 6141320 -------------UGGGCAGCCAGCAGCUGCACCUUUAUCGCUCACACUGGAUCUGGGCGCUCCAGCGACAAACACGAGACGCGCGCCA---GUCUUGGGGAUAUUUUUAAC -------------.(((((((.....))))).))..((((.((((....(((.((((((((((...((.......)).)).)))).)))---))))))))))))........ ( -31.90, z-score = -1.32, R) >consensus _______C_AG__AAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC_______AAUGGGACAAUCCCGAGACGUCUCGCGGAUAUUUUUAAC .......................((....)).(((..(((((......)))))..))).................((((((...........)))))).............. (-12.84 = -12.57 + -0.26)


| Location | 13,618,763 – 13,618,863 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.58094 |
| G+C content | 0.57489 |
| Mean single sequence MFE | -36.99 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13618763 100 + 27905053 -----UCCCAUUGC--GAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUGCUCGGA--------GGAGCUGAGGAGCA-AGGAGCAGGCGAGGAUAA -----..((.((((--...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.))))...))))).-))))))..)))))).... ( -41.70, z-score = -2.72, R) >droSim1.chr3R 19645720 100 - 27517382 -----UCCCAUUGC--GAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------GGAGCAGAGGAGCA-AGGAGCAGGCGAGGAUCA -----..((.((((--...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.))))...))))).-))))))..)))))).... ( -43.20, z-score = -2.97, R) >droSec1.super_12 1587892 100 - 2123299 -----UCCCAUUGC--GAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------GGAGCAGAGGAGCA-AGGAGCAGGCGAGGAUCA -----..((.((((--...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.))))...))))).-))))))..)))))).... ( -43.20, z-score = -2.97, R) >droYak2.chr3R 2197721 109 + 28832112 -----UCCCAUUGC--GAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCAGCUUAACUUGGAUGAACAGAGAAGCAGAGGAGCAGAGUAGCAGGCGAGGAUCA -----((((.((((--...((((((.((((.(((((..((......))..)))))...)))).((((...(((......)))..))))..))))))......))))..).)))... ( -37.30, z-score = -2.72, R) >droEre2.scaffold_4770 9734228 101 - 17746568 -----UCCCAUUGC--GAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUACUUGGA--------GGAGCAGAGGAGCAGAAGAGCAGGCGAAGAUCA -----((((..(((--...((((((.((((.(((((..((......))..)))))...)))).(((((.......--------)))))..))))))......))))).))...... ( -33.20, z-score = -1.29, R) >droAna3.scaffold_13340 12913588 91 - 23697760 UCUCUUUUUUUUGUGCACUGGGCCUAGAUCGAGUUGGCUCGAUAAAGGUGCAGCUGCAG--UGG--AAAGUGGAA--------GGGGCAUCAG-GCA-UGGGGUA----------- .((((((((((..(((((((.((((..((((((....))))))..)))).))).))).)--..)--)))...)))--------)))((..((.-...-))..)).----------- ( -31.90, z-score = -1.83, R) >dp4.chr2 25969182 87 + 30794189 ----------UCGCU-GGAGCGCCCAGAUCCAGUGUGAGCGAUAAAGGUGCAGCUGCUGGCUGCCCAGA---GAAU-------GGGGCAGGGGGGCA-GGGGGCAGACA------- ----------..(((-...((.(((....(((((((..(((.......))).)).)))))((((((...---....-------.))))))))).)).-...))).....------- ( -32.40, z-score = 0.50, R) >droPer1.super_6 1302745 87 + 6141320 ----------UCGCU-GGAGCGCCCAGAUCCAGUGUGAGCGAUAAAGGUGCAGCUGCUGGCUGCCCAGA---GAAU-------GGGGCGGGGGGGCA-GGGGGCAGGCA------- ----------..(((-...((.(((....(((((((..(((.......))).)).)))))((((((...---....-------.)))))).......-))).)).))).------- ( -33.00, z-score = 0.92, R) >consensus _____UCCCAUUGC__GAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUAACUCGGA________GGAGCAGAGGAGCA_AGGAGCAGGCGAGGAUCA ...................((.(((......(((((..((......))..)))))((...((.(((...................))).))...))..))).))............ (-13.62 = -14.19 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:21 2011