
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,824,277 – 7,824,369 |
| Length | 92 |
| Max. P | 0.974164 |

| Location | 7,824,277 – 7,824,369 |
|---|---|
| Length | 92 |
| Sequences | 15 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.51 |
| Shannon entropy | 0.72913 |
| G+C content | 0.55178 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -12.82 |
| Energy contribution | -12.61 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
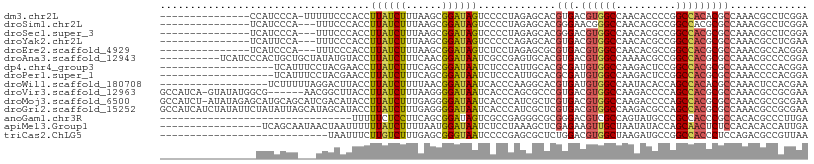
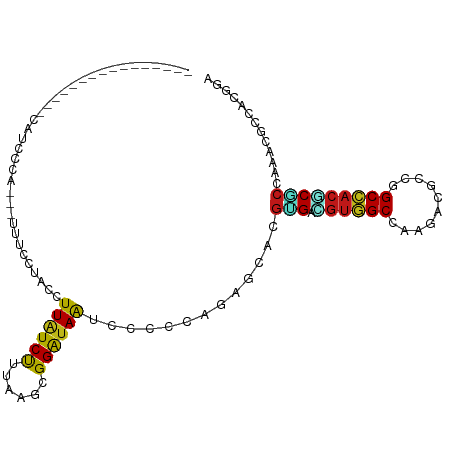
>dm3.chr2L 7824277 92 - 23011544 ---------------CCAUCCCA-UUUUUCCCACCUUAUCUUUAAGCGGAUAGUCCCCUAGAGCACGUGACGUGGCCAACACCCCGGCCACACGCCAAACGCCUCGGA ---------------.....((.-.......(((.....(((((.(.((.....))).)))))...)))..((((((........))))))..............)). ( -20.20, z-score = -0.98, R) >droSim1.chr2L 7619984 90 - 22036055 ---------------UCAUCCCA---UUUCCCACCUUAUCUUUAAGCGGAUAGUCCCCUAGAGCACGGGAACGGGCCAACACGCCGGCCACGCGCCAAACGCCUCGGA ---------------........---..(((...((((....)))).)))......((.((.((...((..((((((........)))).))..))....)))).)). ( -21.30, z-score = 0.82, R) >droSec1.super_3 3338262 90 - 7220098 ---------------UCAUCCCA---UUUCCCACCUUAUCUUUAAGCGGAUAGUCCCCUAGAGCACGGGACGUGGCCAACACGCCGGCCACGCGCCAAACGCCUCGGA ---------------.....((.---..(((...((((....)))).)))..(((((.........)))))((((((........))))))(((.....)))...)). ( -26.60, z-score = -1.27, R) >droYak2.chr2L 17250609 90 + 22324452 ---------------UCAUUCCA---UUUCCCACCUUAUCUUUAAGCGGAUAGUCCCCCAGAGCACGUGACGUGGCCAACACGCCGGCCACGCGCCAAACGCCUCGAA ---------------((......---..(((...((((....)))).)))..........((((..(((.(((((((........)))))))))).....).))))). ( -21.60, z-score = -1.15, R) >droEre2.scaffold_4929 16745256 90 - 26641161 ---------------UCAUCCCA---UUUCCCACCUUAUCUUUAAGCGGAUAGUCUCCUAGAGCGCGUGACGUGGCCAACACGCCGGCCACGCGCCAAACGCCACGGA ---------------.....((.---..(((...((((....)))).)))..........(.(((.(((.(((((((........))))))))))....))))..)). ( -25.20, z-score = -1.08, R) >droAna3.scaffold_12943 3555646 98 + 5039921 ----------UCAUCCCACUGCUGCUAUAUGUACCUUAUCUUUCAACGGAUAAUCGCCGAGUGCACGUGACGUGGCCAAAACGCCGGCCACGCGCCAAACGCCCCGGA ----------........(((..((....((((((((((((......)))))).....).))))).(((.(((((((........)))))))))).....))..))). ( -27.00, z-score = -0.59, R) >dp4.chr4_group3 6217691 89 + 11692001 -------------------UCAUUUCCUACGAACCUUAUCUUUCAGCGGAUAAUCUCCCAUUGCACGCGAUGUGGCCAAGACUCCGGCCACGCGCCAAACCCCACGGA -------------------.....(((........((((((......)))))).............(((.(((((((........))))))))))..........))) ( -22.20, z-score = -1.56, R) >droPer1.super_1 3317230 89 + 10282868 -------------------UCAUUUCCUACGAACCUUAUCUUUCAGCGGAUAAUCUCCCAUUGCACGCGAUGUGGCCAAGACUCCGGCCACGCGCCAAACCCCACGGA -------------------.....(((........((((((......)))))).............(((.(((((((........))))))))))..........))) ( -22.20, z-score = -1.56, R) >droWil1.scaffold_180708 6880773 90 + 12563649 ------------------UCUUUUUAGGACUUACCUUAUCUUUUAACGGAUAAUCACCCAAGGCACGUGAUGUGGCCAAUACACCAGCCACACGCCAAACUCCACGAA ------------------........(((......((((((......)))))).............(((.((((((..........))))))))).....)))..... ( -16.80, z-score = -1.33, R) >droVir3.scaffold_12963 13660435 101 + 20206255 GCCAUCA-GUAUAUGGCG------AACGGCUUACCUUAUCUUUAAGGGGAUAAUCACCCAGCGCCCGUGACGUGGCCAAGACCCCAGCCACGCGCCAAACGCCGCGAA (((((..-....))))).------...(((...(((((....)))))((........))...)))((((..(((((..........)))))(((.....))))))).. ( -31.10, z-score = -0.99, R) >droMoj3.scaffold_6500 6652152 107 + 32352404 GCCAUCU-AUAUAGAGCAUGCAGCAUCGACAUACCUUAUCUUUGAGGGGAUAAUCACCCAUCGCUCGUGACGUGGCCAAGACCCCAGCCACGCGCCAAACGCCGCGAA ((.....-.....((((................(((((....)))))((........))...))))(((.((((((..........)))))))))........))... ( -27.20, z-score = -0.44, R) >droGri2.scaffold_15252 7431343 108 + 17193109 GCCAUCAUCUAUAUUCUAUAUUAGCAUAGCAUACCUUAUCUUUGAGGGGAUAAUCACCCAUCGCUCGUGACGUGGCCAAGACGCCAGCCACGCGCCAAACGCCGCGAA ......((((.(((.((((......)))).)))(((((....))))))))).........((((..(((.(.((((......))))).)))(((.....))).)))). ( -24.70, z-score = -0.09, R) >anoGam1.chr3R 12135588 76 - 53272125 --------------------------------UUUUUCUCCUUCAGCGGAUAGUCGCCGAGGGCGCGGGACGUCGCCAGUAUGCCCGCCACCCGCCACACGCCCUUGA --------------------------------.............((((....))))((((((((((((.((((....).)))))))).....(.....)))))))). ( -25.20, z-score = -0.29, R) >apiMel3.Group1 2411517 91 + 25854376 -----------------UCAGCAAUAACUAAUUUUUUAUCUUUUAAUGGAUAAUCUCCUAAAGCUCGAGAAGUUGCUAAUAUACCAGCAACUCUCCACACACCAUUGA -----------------....((((..........((((((......)))))).............((((.((((((........))))))))))........)))). ( -16.50, z-score = -2.97, R) >triCas2.ChLG5 10914194 80 - 18847211 ----------------------------UAAUUUCUUGUCUUUGAGCGGGUAAUCCCCGAGCGCUCUGGACGUGGCUAAGAUGCCGGCCACCCUCCAGACGCCGUUAA ----------------------------..................((((.....)))).(((.((((((.((((((........))))))..)))))))))...... ( -27.70, z-score = -1.81, R) >consensus ________________CAUCCCA___UUUCCUACCUUAUCUUUAAGCGGAUAAUCCCCCAGAGCACGUGACGUGGCCAAGACGCCGGCCACGCGCCAAACGCCACGGA ...................................((((((......)))))).............(((.((((((..........)))))))))............. (-12.82 = -12.61 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:24 2011