| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,575,316 – 13,575,407 |
| Length | 91 |
| Max. P | 0.667250 |

| Location | 13,575,316 – 13,575,407 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.53 |
| Shannon entropy | 0.47545 |
| G+C content | 0.46870 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.11 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
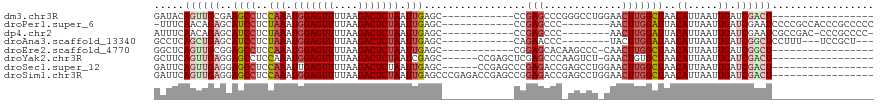
>dm3.chr3R 13575316 91 - 27905053 GAUACAGUUGCGAAGCCUCCAAAUGGAGUUUUAAGACUCUAAUUGAGC------------CCGAGCCCGGGCCUGGAACUUGGCUAACAUUAAUUGAUCGACU----------------- (((.((((((...((((((((..(((((((....))))))).....((------------(((....))))).))))....)))).....)))))))))....----------------- ( -29.40, z-score = -3.22, R) >droPer1.super_6 1265558 99 - 6141320 -UUUCAACAGAGCAUCCUCUAAAUGGAGUUUUAAGACUCUAAUUGAGC------------CCGAGCCC--------AACUUGGAUUACAUUAAUUGAUCGAAUCCCCGCCACCCGCCCCC -........(.((...(((.((.(((((((....))))))).))))).------------(((((...--------..)))))........................))).......... ( -16.00, z-score = -0.88, R) >dp4.chr2 25932002 98 - 30794189 AUUUCAACAGAGCAUCCUCUAAAUGGAGUUUUAAGACUCUAAUUGAGC------------CCGAGCCC--------AACUUGGAUUACAUUAAUUGAUCGAAUCGCCGAC-CCCGCCCC- ...((((.((((....))))...(((((((....))))))).))))((------------(((((...--------..)))))..............(((......))).-...))...- ( -17.60, z-score = -0.91, R) >droAna3.scaffold_13340 12869450 94 + 23697760 GCCUCAGCUGAGCAUCCUCUAAAUGGAGUUUUAAGACUCUAAUUGAGC------------CAGAACCC--------UACUUGGAUAACAUUAAUUGAUCGGCUCCUUU---UCCGCU--- .....(((.(((...........(((((((....)))))))...((((------------(.....((--------.....))....((.....))...)))))...)---)).)))--- ( -19.90, z-score = -0.67, R) >droEre2.scaffold_4770 9690891 90 + 17746568 GGCUCAGUUGCGGAGCCUCCAAAUGGAGUUUUAAGACUCUAAUUGAGC------------CGGAGCACAAGCCC-CAACUUGGCUAACAUUAAUUGAUCGGCU----------------- (((((.......)))))..(((.(((((((....))))))).)))(((------------((((((.((((...-...)))))))..((.....)).))))))----------------- ( -30.90, z-score = -2.51, R) >droYak2.chr3R 2153174 96 - 28832112 GCUUCAGUUGAGGAGCCUCCAAAUGGAGUUUUAAGACUCUAAUCGAGC------CCGAGCUCGAGCCCAAGUCU-GAACUGUGCUAACAUUAAUUGAUCGACU----------------- ...((((((((..(((((((....)))).....(((((....((((((------....)))))).....)))))-.......)))....))))))))......----------------- ( -28.60, z-score = -2.25, R) >droSec1.super_12 1537185 97 + 2123299 GAUUCAGUUGAGGAGCCUCCAAAUUGAGUUUUAAGACUCUAAUUGAGC------CCGAGCCCGAGACCGAGCCUGGAACUUGGCUAACAUUAAUUGAUCGACU----------------- (((.(((((((..((((..(((...(((((....)))))...)))...------(((.((.((....)).)).))).....))))....))))))))))....----------------- ( -26.10, z-score = -2.20, R) >droSim1.chr3R 19600694 103 + 27517382 GAUUCAGUUGAGGAGCCUCCAAAUGGAGUUUUAAGACUCUAAUUGAGCCCGAGACCGAGCCGGAGACCGAGCCUGGAACUUGGCUAACAUUAAUUGAUCGACU----------------- (((.(((((((..((((..(((.(((((((....))))))).))).........(((.(((((...))).)).))).....))))....))))))))))....----------------- ( -31.20, z-score = -2.49, R) >consensus GAUUCAGUUGAGGAGCCUCCAAAUGGAGUUUUAAGACUCUAAUUGAGC____________CCGAGCCC_AGCCU_GAACUUGGCUAACAUUAAUUGAUCGACU_________________ ...((((.(((..((((..(((.(((((((....))))))).))).................(((.............)))))))....))).))))....................... (-10.75 = -11.11 + 0.36)
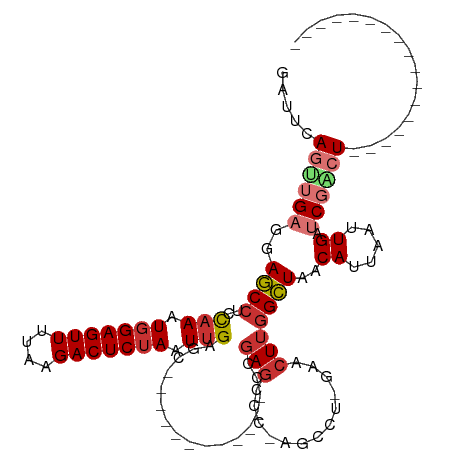

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:15 2011