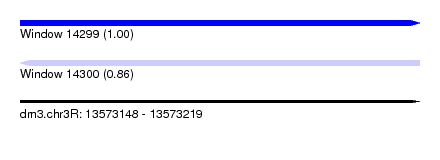
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,573,148 – 13,573,219 |
| Length | 71 |
| Max. P | 0.997567 |

| Location | 13,573,148 – 13,573,219 |
|---|---|
| Length | 71 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.54648 |
| G+C content | 0.45115 |
| Mean single sequence MFE | -20.01 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.93 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
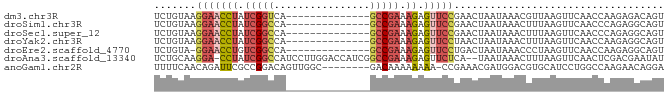
>dm3.chr3R 13573148 71 + 27905053 UCUGUAAGGAACCUAUCGGUCA--------------GCCGAAAGAGUUCCGAACUAAUAAACGUUAAGUUCAACCAAGAGACAGU .((((..(((((((.((((...--------------.)))).)).)))))((((((((....))).))))).........)))). ( -22.50, z-score = -4.51, R) >droSim1.chr3R 19598525 71 - 27517382 UCUGUAAGGAACCUAUCGGCCA--------------GCCGAAAGAGUUCCGAACUAAUAAACUUUAAGUUCAACCCAGAGGCAGU .((((..(((((((.((((...--------------.)))).)).)))))(((((((......)).))))).........)))). ( -22.60, z-score = -3.98, R) >droSec1.super_12 1535168 71 - 2123299 UCUGUAAGGAACCUAUCGGCCA--------------GCCGAAAGAGUUCCGAACUAAUAAACUUUAAGUUCAACCCAGAGGCAGU .((((..(((((((.((((...--------------.)))).)).)))))(((((((......)).))))).........)))). ( -22.60, z-score = -3.98, R) >droYak2.chr3R 2150995 71 + 28832112 UCUGUAAGGAACCUAUCGGCCA--------------GCCGAAAGAGUUCCUAACUAAUAAACUUUAAGUUCAACCAAGAGGCAGU .((((.((((((((.((((...--------------.)))).)).))))))..........((((..(.....)..)))))))). ( -19.80, z-score = -3.09, R) >droEre2.scaffold_4770 9688892 70 - 17746568 UCUGUA-GGAACCUGUCGGCCA--------------GCCGAAAGAGUUCCUGACUAAUAAACCCUAAGUUCAACCAAGAGGCAGU .(((((-(((((((.((((...--------------.)))).)).)))))).................(((......))))))). ( -19.10, z-score = -2.14, R) >droAna3.scaffold_13340 12867459 82 - 23697760 UCUGCAAGGA-CCUAUCGGCCAUCCUUGGACCAUCGGCCGAAAGAGUUCUCA--UAAUAAACUUUAAGUUCAACUCGACGAAUAU ......((((-(((.((((((.((....)).....)))))).)).)))))..--.............((((........)))).. ( -17.80, z-score = -1.16, R) >anoGam1.chr2R 50032596 76 + 62725911 UUUUCAACAGAUUCGCCGGACAGUUGGC--------GACAAAAAAAA-CCGAAACGAUGGACGUGCAUCCUGGCCAAGAACAGGA ..............((((..((.((((.--------...........-)))).....))..)).)).(((((........))))) ( -15.70, z-score = 0.11, R) >consensus UCUGUAAGGAACCUAUCGGCCA______________GCCGAAAGAGUUCCGAACUAAUAAACUUUAAGUUCAACCAAGAGGCAGU .......(((((((.(((((................))))).)).)))))................................... (-10.20 = -10.93 + 0.74)
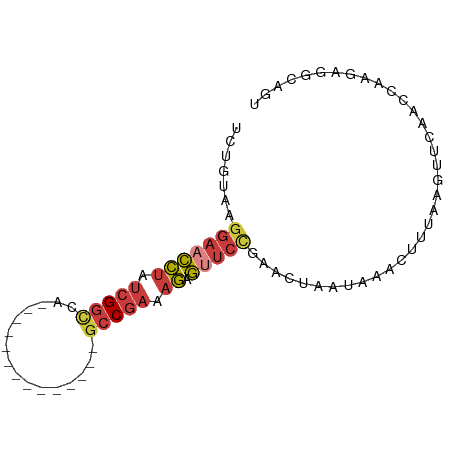

| Location | 13,573,148 – 13,573,219 |
|---|---|
| Length | 71 |
| Sequences | 7 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.54648 |
| G+C content | 0.45115 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
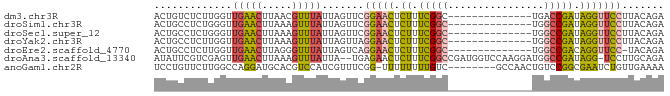
>dm3.chr3R 13573148 71 - 27905053 ACUGUCUCUUGGUUGAACUUAACGUUUAUUAGUUCGGAACUCUUUCGGC--------------UGACCGAUAGGUUCCUUACAGA .((((.........(((((...........)))))(((((.((.((((.--------------...)))).)))))))..)))). ( -20.70, z-score = -2.49, R) >droSim1.chr3R 19598525 71 + 27517382 ACUGCCUCUGGGUUGAACUUAAAGUUUAUUAGUUCGGAACUCUUUCGGC--------------UGGCCGAUAGGUUCCUUACAGA ......((((....(((((...........)))))(((((.((.((((.--------------...)))).)))))))...)))) ( -22.20, z-score = -2.06, R) >droSec1.super_12 1535168 71 + 2123299 ACUGCCUCUGGGUUGAACUUAAAGUUUAUUAGUUCGGAACUCUUUCGGC--------------UGGCCGAUAGGUUCCUUACAGA ......((((....(((((...........)))))(((((.((.((((.--------------...)))).)))))))...)))) ( -22.20, z-score = -2.06, R) >droYak2.chr3R 2150995 71 - 28832112 ACUGCCUCUUGGUUGAACUUAAAGUUUAUUAGUUAGGAACUCUUUCGGC--------------UGGCCGAUAGGUUCCUUACAGA ...(((....)))(((((.....)))))...((.((((((.((.((((.--------------...)))).)))))))).))... ( -22.00, z-score = -2.34, R) >droEre2.scaffold_4770 9688892 70 + 17746568 ACUGCCUCUUGGUUGAACUUAGGGUUUAUUAGUCAGGAACUCUUUCGGC--------------UGGCCGACAGGUUCC-UACAGA .(((.((((.((.....)).))))..........((((((.((.((((.--------------...)))).)))))))-).))). ( -21.60, z-score = -1.20, R) >droAna3.scaffold_13340 12867459 82 + 23697760 AUAUUCGUCGAGUUGAACUUAAAGUUUAUUA--UGAGAACUCUUUCGGCCGAUGGUCCAAGGAUGGCCGAUAGG-UCCUUGCAGA ....((((((.((((((.....(((((....--...)))))..))))))))))))..(((((...(((....))-)))))).... ( -20.40, z-score = -0.41, R) >anoGam1.chr2R 50032596 76 - 62725911 UCCUGUUCUUGGCCAGGAUGCACGUCCAUCGUUUCGG-UUUUUUUUGUC--------GCCAACUGUCCGGCGAAUCUGUUGAAAA (((((........))))).............((((..-(........((--------(((........)))))....)..)))). ( -14.80, z-score = 0.79, R) >consensus ACUGCCUCUUGGUUGAACUUAAAGUUUAUUAGUUCGGAACUCUUUCGGC______________UGGCCGAUAGGUUCCUUACAGA .............(((((.....)))))...((..(((((.((.(((((................))))).)))))))..))... (-10.76 = -10.98 + 0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:12 2011