| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,561,292 – 13,561,385 |
| Length | 93 |
| Max. P | 0.983350 |

| Location | 13,561,292 – 13,561,385 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Shannon entropy | 0.35440 |
| G+C content | 0.35974 |
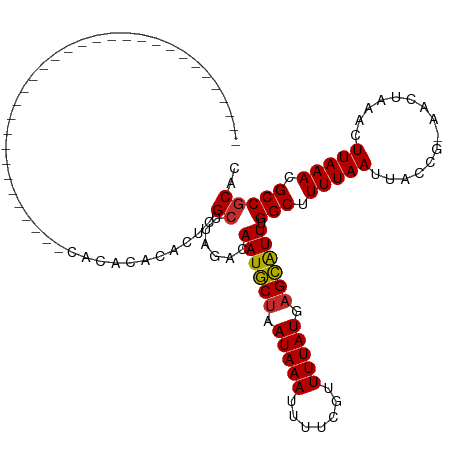
| Mean single sequence MFE | -14.98 |
| Consensus MFE | -13.11 |
| Energy contribution | -13.04 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
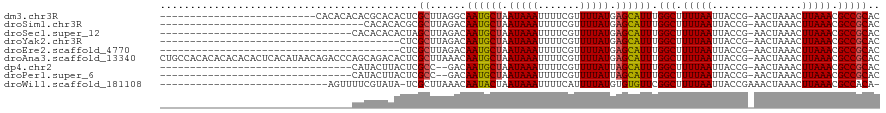
>dm3.chr3R 13561292 93 - 27905053 --------------------------CACACACACGCACACUCGCUUAGGCAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC --------------------------.........((......((....))((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -15.94, z-score = -1.47, R) >droSim1.chr3R 19586358 85 + 27517382 ----------------------------------CACACACGCGCUUAGACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC ----------------------------------.......((........((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -14.74, z-score = -1.64, R) >droSec1.super_12 1522150 87 + 2123299 --------------------------------CACACACACUAGCUUAGACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC --------------------------------...........((......((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -14.14, z-score = -1.95, R) >droYak2.chr3R 2139272 79 - 28832112 ----------------------------------------CUCGCUUAGACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC ----------------------------------------...((......((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -14.24, z-score = -1.79, R) >droEre2.scaffold_4770 9677478 79 + 17746568 ----------------------------------------CUCGCUUAGACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC ----------------------------------------...((......((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -14.24, z-score = -1.79, R) >droAna3.scaffold_13340 12856948 119 + 23697760 CUGCCACACACACACACUCACAUAACAGACCCAGCAGACACUCGCUUAAACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC ((((.............................))))......((......((((((.(((((.......))))).)))))).(((.(((((......-........))))).))))).. ( -16.29, z-score = -1.56, R) >dp4.chr2 25922298 85 - 30794189 --------------------------------CAUACUUACUCGCC--GACAAUGCUAAUAAAUUUUCGUUUUAUUAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC --------------------------------...........((.--...((((((((((((.......)))))))))))).(((.(((((......-........))))).))))).. ( -17.74, z-score = -3.67, R) >droPer1.super_6 1255893 85 - 6141320 --------------------------------CAUACUUACUCGCC--GACAAUGCUAAUAAAUUUUCGUUUUAUUAGCAUUUGGCUUUUAAUUACCG-AACUAAACUUAAACGCCGCAC --------------------------------...........((.--...((((((((((((.......)))))))))))).(((.(((((......-........))))).))))).. ( -17.74, z-score = -3.67, R) >droWil1.scaffold_181108 1569286 90 - 4707319 ----------------------------AGUUUUCGUAUA-UCGCUUAAACAAUACUAAUAAAUUUUCAUUUUAUGUGUGUUCGGCUUUUAAUUACCGAAACUAAACUUAAACGCCACA- ----------------------------.((((..((...-..))..))))(((((..(((((.......)))))..))))).(((.(((((...............))))).)))...- ( -9.76, z-score = -0.00, R) >consensus ________________________________CACACACACUCGCUUAGACAAUGCUAAUAAAUUUUCGUUUUAUGAGCAUUUGGCUUUUAAUUACCG_AACUAAACUUAAACGCCGCAC ...........................................((......((((((.(((((.......))))).)))))).(((.(((((...............))))).))))).. (-13.11 = -13.04 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:10 2011