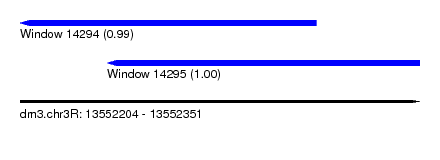
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,552,204 – 13,552,351 |
| Length | 147 |
| Max. P | 0.997461 |

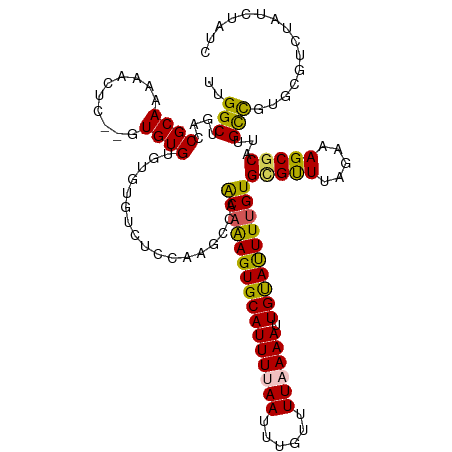
| Location | 13,552,204 – 13,552,313 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Shannon entropy | 0.40150 |
| G+C content | 0.35907 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -14.68 |
| Energy contribution | -16.36 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13552204 109 - 27905053 GCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGUAUUUUGUGCGUUUAGAAAGCGCAUGGCCGU--UUCUAUCUAUCCAGUCAAAG--------AAAUCUCUCUGAUUUUUGAAGUC ((((((((((((((((((((......)))))).))))))))))(((((.....))))).))))...--..............(((..(--------(((((.....))))))))).... ( -27.20, z-score = -3.31, R) >droEre2.scaffold_4770 9667871 102 + 17746568 ---------GUGCAUUUUAAUUUGUUUU-AAAUUGUAUUUUGUGUGUUUAGACAGCGCAGUGC-------CUGUGCAUACAGUCAAAGACACAAAGAAAUCUCUCUGAAUUUCAGAGUC ---------(((((.(((((......))-))).)))))(((((((.(((.(((.((((((...-------)))))).....)))))).))))))).......(((((.....))))).. ( -28.80, z-score = -2.78, R) >droYak2.chr3R 2129898 112 - 28832112 GCCAACAGAGUGCAUUUUAAUUGGUUUU-AAAUUGUAUUUUGUGCGCUUAGAAAGCUCAUUGUC------UUGUUUAUUGAGUCAAAGACACAAUGAAAUUUCUCUGAAUUUUUAGGUC ((..((((((((((.(((((......))-))).))))))))))))(((((((((..(((..(..------...(((((((.(((...))).)))))))....)..))).))))))))). ( -29.70, z-score = -2.86, R) >droSec1.super_12 1513005 119 + 2123299 GCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGCACUUUGUGCGUUUAGAAAGCGCAUUGCUGUGCGUCUAUCUAUCCGGACAAAGAUGCCCAGAAAUCUCUCCGAUUUUUGAAGUC .((.((((((((((((((((......)))))).))))))))))..(..((((.(((((((....))))).)).))))..))).....(((...((((((((.....))))))))..))) ( -38.10, z-score = -4.66, R) >droSim1.chr3R 19577134 119 + 27517382 GCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGCAUUUUGUGCGUUUAGAAAGCGCAUUGCUGUGCGUCUAUCUAUCCGGACAAAGAUGCCCACAAAUCUCUCCGAUUUUUGAAGUC ((..((((((((((((((((......)))))).))))))))))))(..((((.(((((((....))))).)).))))..)(((...((((........)))).)))............. ( -32.90, z-score = -3.25, R) >consensus GCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGUAUUUUGUGCGUUUAGAAAGCGCAUUGCCGU___UCUAUCUAUCCAGUCAAAGACACAAAGAAAUCUCUCUGAUUUUUGAAGUC ....((((((((((((((((......)))))).))))))))))(((((.....))))).............................(((.....((((((.....))))))....))) (-14.68 = -16.36 + 1.68)

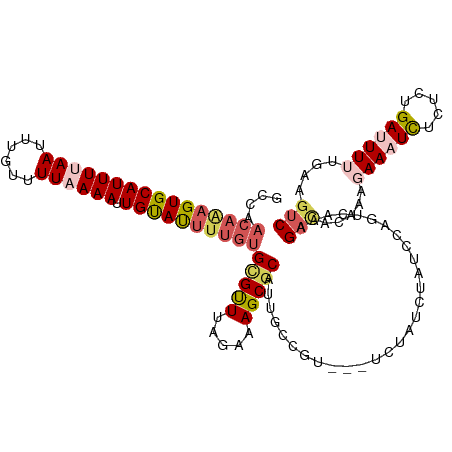
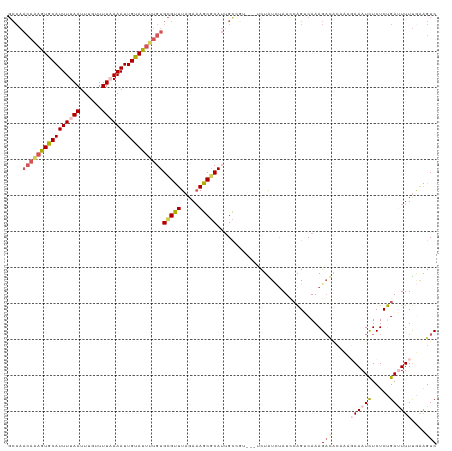
| Location | 13,552,236 – 13,552,351 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Shannon entropy | 0.37770 |
| G+C content | 0.39569 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13552236 115 - 27905053 UUGGCUGACCGCAAAAACUC--GUGUGUGUGUGUCUCCAAGCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGUAUUUUGUGCGUUUAGAAAGCGCAUGGCCGUU--UCUAUCUAUC ((((..(((((((...((..--..)).)))).))).))))((((((((((((((((((((......)))))).))))))))))(((((.....))))).))))....--.......... ( -35.10, z-score = -3.94, R) >droEre2.scaffold_4770 9667911 92 + 17746568 --GGCUGGCCGCAAAAACUC--UUGUGU---------------UGCUAAGUGCAUUUUAAUUUGUUUU-AAAUUGUAUUUUGUGUGUUUAGACAGCGCAGUGCC-UGUGCAUA------ --((....))(((.......--((((((---------------(((((((..(((..(((((((...)-))))))......)))..))))).))))))))....-..)))...------ ( -24.32, z-score = -0.74, R) >droYak2.chr3R 2129938 102 - 28832112 --GGCUGGCCGCAAACACUC--GUGUGU------UUCUAAGCCAACAGAGUGCAUUUUAAUUGGUUUU-AAAUUGUAUUUUGUGCGCUUAGAAAGCUCAUUGUCUUGUUUAUU------ --((....))(((((((...--(((.((------((((((((((.(((((((((.(((((......))-))).))))))))))).)))))).)))).)))))).)))).....------ ( -30.70, z-score = -2.90, R) >droSec1.super_12 1513045 119 + 2123299 UUGGCUGACCGCAAAAACUCGAGUGUGUGUGUGUCUCCAAGCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGCACUUUGUGCGUUUAGAAAGCGCAUUGCUGUGCGUCUAUCUAUC .((((((((((((...((....))...)))).)))....)))))((((((((((((((((......)))))).)))))))))).....((((.(((((((....))))).)).)))).. ( -41.50, z-score = -4.64, R) >droSim1.chr3R 19577174 117 + 27517382 UUGGCUGACCGCAAAAACUC--GUGUGUGUGUGUCUCCAAGCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGCAUUUUGUGCGUUUAGAAAGCGCAUUGCUGUGCGUCUAUCUAUC .((((((((((((...((..--..)).)))).)))....)))))((((((((((((((((......)))))).)))))))))).....((((.(((((((....))))).)).)))).. ( -38.80, z-score = -4.34, R) >consensus UUGGCUGACCGCAAAAACUC__GUGUGUGUGUGUCUCCAAGCCAACAAAGUGCAUUUUAAUUUGUUUUAAAAUUGUAUUUUGUGCGUUUAGAAAGCGCAUUGCCGUGCGUCUAUCUAUC ....((((.(((....((......))..................((((((((((((((((......)))))).))))))))))))).))))...((((((....))))))......... (-20.08 = -20.72 + 0.64)

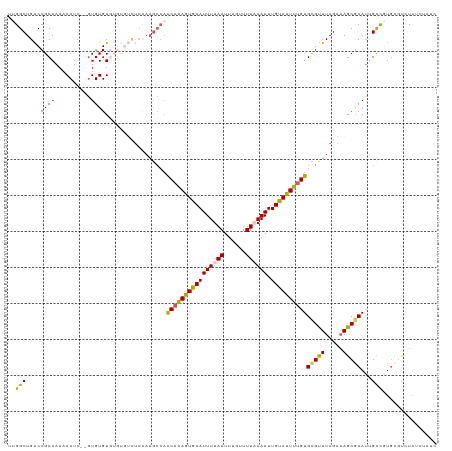
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:08 2011