
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,545,050 – 13,545,165 |
| Length | 115 |
| Max. P | 0.814997 |

| Location | 13,545,050 – 13,545,165 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.45 |
| Shannon entropy | 0.47523 |
| G+C content | 0.36903 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -17.05 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
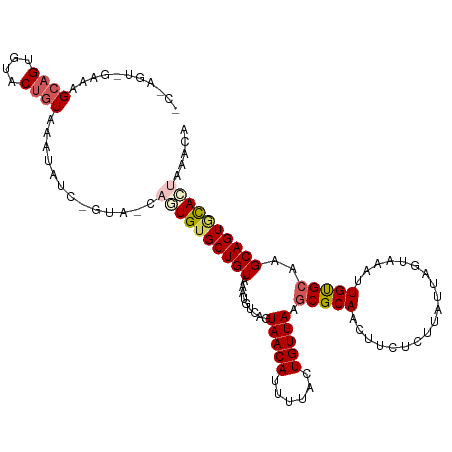
>dm3.chr3R 13545050 115 - 27905053 GCAAGUCGAAAGCAGUGUACUGUAAUUCUC-GUA-CG-UGUGCUGUUAAUGUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGCAAAUUGUGCAAGCAGUUCAAAAAAUA ((........(((((..(((.(((......-.))-))-))..))))).....((((((....))))))....((((((.((((......)).)).))))))..))............. ( -23.60, z-score = 0.02, R) >droEre2.scaffold_4770 9660542 108 + 17746568 -------GGAAGCGGUUUUCAAUGUGCUGAAUUAUCAUUGUGCUGUUAACGUUCCUAACAUUUUACUGUUAAGCGCAACUUCUGUUAUUGGUAUUAUGCGCAAGCAGUACAAAUU--- -------......(((.((((......))))..))).((((((((((........(((((......))))).(((((....((......)).....))))).))))))))))...--- ( -24.70, z-score = -0.48, R) >droYak2.chr3R 2122404 96 - 28832112 -------ACAAGCAGUGUACUGUAAGUACCAGUA-CAUUGUGCUGUAAAUGUUACUAACAUUUUACUGUUAAACGCAACUUC--------------UGCGCAAGCAGUGCACUAAACA -------(((.(((((((((((.......)))))-))))))..)))...((((..(((((......)))))...(((....(--------------(((....)))))))....)))) ( -29.40, z-score = -1.81, R) >droSec1.super_12 1505764 116 + 2123299 CCUAGUUGAAAGCAGUGUACUGUAAAUAUC-GUA-CAGUGUGCUGUAAAUAUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGUACUAAGCA ...(((((...((((..(((((((......-.))-)))))..))))...((.((((((....)))))).))....))))).......((((((.(((((....))))).))))))... ( -32.40, z-score = -2.18, R) >droSim1.chr3R 19569703 116 + 27517382 GCUAGUCGAAAGCAGUGUACUGUAAAUAUC-GUA-CAGUGUGCUGUAAAUAUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGCACUAAGCA (((.(.(....))((((((((((.......-(((-.(((((((((.......)))).))))).)))......((((((.((..((....))..))))))))..)))))))))).))). ( -35.50, z-score = -2.62, R) >consensus _C_AGU_GAAAGCAGUGUACUGUAAAUAUC_GUA_CAGUGUGCUGUAAAUGUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGCACUAAACA ...........((((....)))).............((((((((((.........(((((......))))).((((((...((......))....))))))..))))))))))..... (-17.05 = -18.50 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:06 2011