
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,821,476 – 7,821,557 |
| Length | 81 |
| Max. P | 0.777340 |

| Location | 7,821,476 – 7,821,557 |
|---|---|
| Length | 81 |
| Sequences | 8 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 60.91 |
| Shannon entropy | 0.80812 |
| G+C content | 0.43746 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -3.09 |
| Energy contribution | -4.62 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.13 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7821476 81 - 23011544 GAUUUCAGCUGAAAUUGAU--GAACUUUUGAAAUAUCGAUAGGGGCCGAAGCGAUAACUCUUAUCG---AUAGCCAGCAGGGGUAC ((((((....))))))...--..(((((((...((((((((((((............)))))))))---))).....))))))).. ( -22.40, z-score = -2.26, R) >droAna3.scaffold_12943 3553087 70 + 5039921 ----------GAUUUCAGU--GAGUGACGGCACUAUCGGCGGGUGCCAGUGCGAUA-CCGGUAUCG---AUAGUUCCCGCAGAUAC ----------.........--.((((....))))(((.(((((.((.....(((((-....)))))---...)).))))).))).. ( -20.90, z-score = -0.41, R) >droEre2.scaffold_4929 16742471 75 - 26641161 GAUUUCAGCUGAAAUUUAU--GACCUUUUGAUAUAUCGAUAGGG------GCGAUAGCUCUAAUCG---AUAGUCAGCAGAGGUAC ((((((....))))))...--.((((((((((.(((((((.(((------((....))))).))))---)))))))..)))))).. ( -26.10, z-score = -4.21, R) >droYak2.chr2L 17247770 81 + 22324452 GAUUUCAGCUGAAAUUUAU--GACCUAUUGAUAUAUCGAUAGGGACCGCUGCGAUAACUGUUAUCG---AUAAUCAGCAGAGGUAC .(((((.(((((.......--..((((((((....))))))))........(((((.....)))))---....))))).))))).. ( -23.00, z-score = -3.00, R) >droSec1.super_3 3335473 81 - 7220098 GAUUUCAGCUAAAAUUGAU--GACCUAUUGAAAUAUCGAUAGGGGCCGAAACGAUAACUCUUAUCG---AUAGCCAGCAGGGGUAC .((..(.(((.........--..((((((((....))))))))(((.....(((((.....)))))---...)))))).)..)).. ( -22.20, z-score = -2.66, R) >droSim1.chr2L 7617203 81 - 22036055 GAUUUCAGCUGAAAUUGAU--GACCUACUGAAAUAUCGAUAGGGGCCGAAGCGAUAACUCUUAUCG---AUAGCCAGCAGGGGUAC ((((((....))))))...--..(((.(((...((((((((((((............)))))))))---)))..))).)))..... ( -24.10, z-score = -2.60, R) >droWil1.scaffold_180708 6876496 77 + 12563649 --UUUCAGCUAGAGCUGCA--AAUCCAUCG----AUUGA-GUGCGAUAAAUCGAUAUUUCUUAUCGCUCAUAUCGAGCGGAACCAC --...((((....))))..--..(((.(((----(((((-..(((((((...((....)))))))))))).)))))..)))..... ( -21.40, z-score = -2.17, R) >droGri2.scaffold_15252 7429218 78 + 17193109 CUAUUCAGUCGUAUUUGGCUGAAUAGAUUAGUAUCCCGUUUAGGG----ACCUGCAACUGUAAGC----ACUGCUGUAUGAAAUAC (((((((((((....)))))))))))..(((((((((.....)))----)..(((........))----).))))).......... ( -23.90, z-score = -3.05, R) >consensus GAUUUCAGCUGAAAUUGAU__GACCUAUUGAAAUAUCGAUAGGGGCCGAAGCGAUAACUCUUAUCG___AUAGCCAGCAGAGGUAC ...(((.(((.............(((.((((....)))).)))(((.....(((((.....)))))......)))))).))).... ( -3.09 = -4.62 + 1.53)


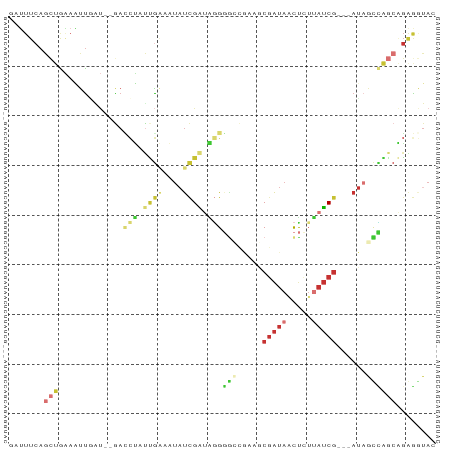
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:23 2011