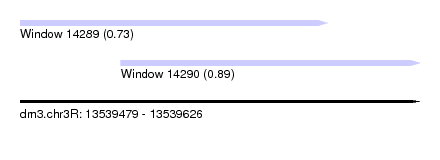
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,539,479 – 13,539,626 |
| Length | 147 |
| Max. P | 0.886549 |

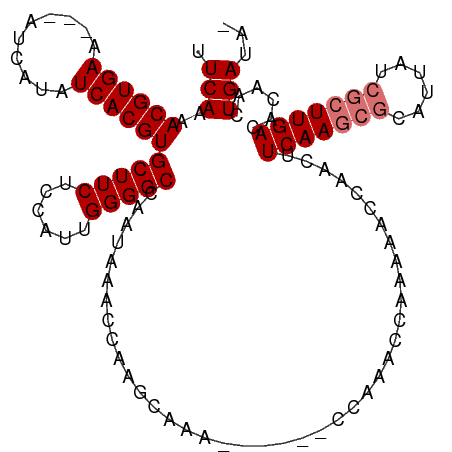
| Location | 13,539,479 – 13,539,592 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.37639 |
| G+C content | 0.40639 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734329 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 13539479 113 + 27905053 ACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGAAAAAAUCAUAUCACGUGCUUCUCCAUUGGGGCCAACAAACCAAGCAAA------CCAAACCAAACACCAA .(((((..........((((((.(((...))).))))))....((((((..........))))))..........))))).................------................ ( -18.00, z-score = -1.64, R) >droSim1.chr3R 19564094 110 - 27517382 UCCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA------CCAAACCAAAAACCAA .(((((..........((((((.(((...))).))))))....((((((---.......))))))..........))))).................------................ ( -18.20, z-score = -1.93, R) >droSec1.super_12 1500224 110 - 2123299 ACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA------CCAAACCAAAAACCAA .(((((..........((((((.(((...))).))))))....((((((---.......))))))..........))))).................------................ ( -18.60, z-score = -1.94, R) >droYak2.chr3R 2115847 116 + 28832112 ACCCCACUAAUGAAACAAUUAACCGGUUUAUAUUUAAUUCAAAACGUGA---AAUCACAUCACGUGCUUCUCCUUUGGGGCCAAUAAACCAGGCAAAAAAAAACCAAACCAAAAACCAA ...((..((((......))))...(((((((............((((((---.......))))))(((((......)))))..))))))).)).......................... ( -19.70, z-score = -1.61, R) >droAna3.scaffold_12911 3255130 89 - 5364042 GCCUCGCCAAUAAAACAAUUAACCGGUUUCCGUUUAAAUGGAAACGUGA---AGUCGCAUCACGUGCACCGAGCGGGCCGGCCAUCAACCAA--------------------------- (((.(((.(((......)))...((((((((((....))))))((((((---.......))))))..)))).)))))).((.......))..--------------------------- ( -22.70, z-score = -0.41, R) >consensus ACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA___AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA______CCAAACCAAAAACCAA .((((...........((((((.((.....)).))))))....((((((..........))))))...........))))....................................... (-13.00 = -13.64 + 0.64)

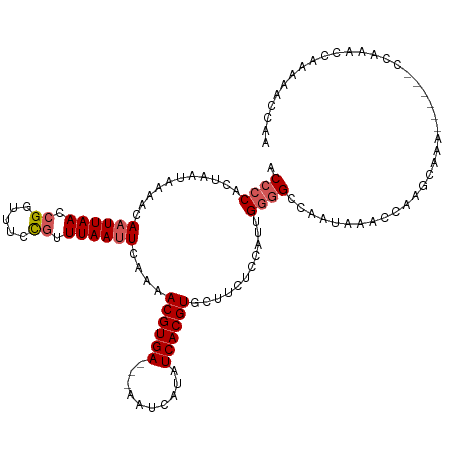
| Location | 13,539,516 – 13,539,626 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Shannon entropy | 0.14581 |
| G+C content | 0.40460 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -16.45 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13539516 110 + 27905053 UUCAAAACGUGAAAAAAUCAUAUCACGUGCUUCUCCAUUGGGGCCAACAAACCAAGCAAA------CCAAACCAAACACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUAG .(((..((((((..........))))))(((((......)))))................------..................(((((((......)))))))......)))... ( -19.30, z-score = -2.07, R) >droSim1.chr3R 19564131 106 - 27517382 UUCAAAACGUGAA---AUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA------CCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUA- .(((..((((((.---......))))))((((....((((....)))).....))))...------..................(((((((......)))))))......)))..- ( -20.20, z-score = -2.35, R) >droSec1.super_12 1500261 106 - 2123299 UUCAAAACGUGAA---AUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA------CCAAACCAAAAACCAACUUCAAGCGCUUUAUCGCUUGACACAACUGAUA- .(((..((((((.---......))))))((((....((((....)))).....))))...------..................(((((((......)))))))......)))..- ( -20.20, z-score = -2.35, R) >droYak2.chr3R 2115884 108 + 28832112 UUCAAAACGUGAA---AUCACAUCACGUGCUUCUCCUUUGGGGCCAAUAAACCAGGCAAAAAAAAACCAAACCAAAAACCAACUUCAA----CUUAUCGCUUGACACAACUGAUA- .(((..((((((.---......))))))(((((......)))))........(((((...............................----......))))).......)))..- ( -14.49, z-score = -0.60, R) >consensus UUCAAAACGUGAA___AUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCAAA______CCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUA_ .(((..((((((..........))))))(((((......)))))........................................(((((((......)))))))......)))... (-16.45 = -17.20 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:04 2011