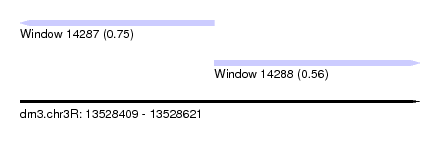
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,528,409 – 13,528,621 |
| Length | 212 |
| Max. P | 0.745785 |

| Location | 13,528,409 – 13,528,512 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 62.92 |
| Shannon entropy | 0.64636 |
| G+C content | 0.34945 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -9.20 |
| Energy contribution | -12.20 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
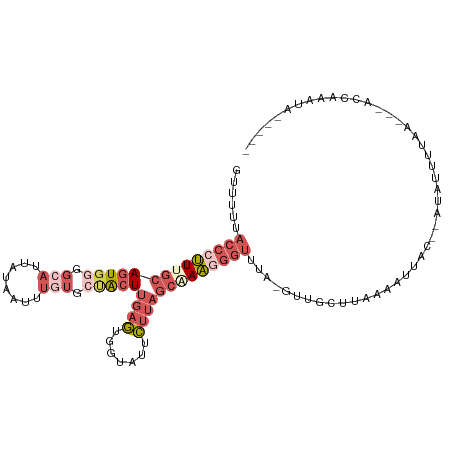
>dm3.chr3R 13528409 103 - 27905053 GUUUUCACCCUUUGCAGUGGGGCAUUAUAAUUUGUGCUACUUGAGUGGUAUUCUUAGCAAAAGGUUUA-GUUGCUUUAAAAUACUAAUACUUUUA---ACUAGAUAA---- ......((((((...(((((.(((........))).))))).))).)))....((((..(((((((((-((...........))))).)))))).---.))))....---- ( -21.00, z-score = -0.32, R) >droSec1.super_12 1489134 82 + 2123299 GUUUUUACCCUUUGCAGUGGGGCAUUAUAAUUUGUGCUACUUGAGUGGUAUUCUUAGCAAAGGGUUUAUUUUAACUAGAAGC----------------------------- ((((((((((((((((((((.(((........))).)))))((((.......)))))))))))))...........))))))----------------------------- ( -24.30, z-score = -2.68, R) >droYak2.chr3R 2101104 103 - 28832112 GUUUUUACCCUUUGCAGUGGGGCAUUAUAAUUUGUGCUACUUGAGGGGUAUUCUUAGCUAAGGGUUUA-GUUGGUUAAAAUUAC--AUGUUUUAACGCACCAACUA----- (((..(((((((((.(((((.(((........))).)))))))))))))).....)))........((-((((((((((((...--..))))))....))))))))----- ( -30.20, z-score = -2.50, R) >droEre2.scaffold_4770 9643817 108 + 17746568 GUUUUUACCCUUUGCAGUGGGGCAUUAUAAUUUUUGACACUUGAGUGGUAUUCUUAGCUAAGGGUUUA-GUUGGUUAAAAUUAC--AUGUUUUAACGUACCAACUAGGAGA ......((((((.((((((...((..(....)..)).))))((((.......)))))).))))))(((-((((((((((((...--..))))))....))))))))).... ( -25.50, z-score = -1.05, R) >droAna3.scaffold_12911 3246109 97 + 5364042 -UUUUAUACACUUGCAGU-GGGUAUUAUAAUUCUGAAUGGUCG-UUGGCAACGGAAAAGAGAACUUUA-AUGACCGUGAGUCGUAAAGAAAACUA---AUUAAA------- -((((.((((((..(...-((((......)))).....(((((-(((....)...((((....)))))-)))))))..))).))).)))).....---......------- ( -18.50, z-score = -0.75, R) >consensus GUUUUUACCCUUUGCAGUGGGGCAUUAUAAUUUGUGCUACUUGAGUGGUAUUCUUAGCAAAGGGUUUA_GUUGCUUAAAAUUAC__AUAUUUUAA___ACCAAAUA_____ ......((((((((((((((.(((........))).)))))((((.......))))))))))))).............................................. ( -9.20 = -12.20 + 3.00)

| Location | 13,528,512 – 13,528,621 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Shannon entropy | 0.30327 |
| G+C content | 0.46386 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -11.73 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
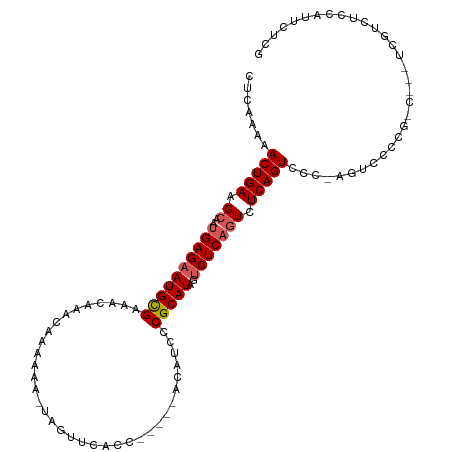
>dm3.chr3R 13528512 109 + 27905053 CUCAAAAACUGAAGCAAUGAGAAUGCGAAACAAACAAAAAAAUAGUUCACCAC--CACAUCCCGCAUAGUCUCAGUCUCAGUCCC-AGUACCCGCCG--UCGUCUCCAUUCUCG .......(((((..(..((((((((((..........................--.......)))))..))))))..)))))...-...........--............... ( -14.51, z-score = -1.64, R) >droSim1.chr3R 19556621 101 - 27517382 CUCAAAAACUGAAGCAA-GAGAAUGCGAA-CAAACAAAAAU-AGUUUCACC-----ACAUCCCGCAUAGUCUCAGUCUCAGUCCCAGGUGCCCG-----UCGUUUCCAUUCUCG .................-(((((((.(((-(((((......-.))))....-----.......((((.(.((.......)).)....))))...-----..)))).))))))). ( -15.20, z-score = -0.87, R) >droSec1.super_12 1489216 103 - 2123299 CUCAAAAACUGAAGCAAUGAGAAUGCGAAACAAACAAAAAA-UAGUUCACC-----ACAUCCCGCAUAGUCUCAGUCUCAGUCCCCAGUGCCCG-----UCGUUUCCAUUCUGG .......(((((..(..((((((((((..............-.........-----......)))))..))))))..)))))..((((......-----...........)))) ( -16.88, z-score = -1.39, R) >droYak2.chr3R 2101207 112 + 28832112 CUCAAAAACUGAAGCAGUGAGAAUGCGAAACAAACAACAAA-UAGUUUACCACACCAUGUCCCGCAUAGCCUCAGUCUCAGUCCC-AGUCCACGUCUCCGCGUCUCCAUUCUUG .......(((((..(..((((.(((((..((((((......-..)))..........)))..)))))...)))))..)))))...-.....((((....))))........... ( -16.30, z-score = -0.62, R) >droEre2.scaffold_4770 9643925 105 - 17746568 CUCAAAAACUGAAGCACUGAGAAUGUGAAAUAAACAACAAA-UAGUUCACC-----ACAUCCCGCAUAGUCUCAUUCUCAGUCUC-GGUCUCAAUCC--CCGUCUCCAUUCCCG .......(((((.(.((((((((((.((.............-..(....).-----.............)).)))))))))))))-)))........--............... ( -17.55, z-score = -2.70, R) >consensus CUCAAAAACUGAAGCAAUGAGAAUGCGAAACAAACAAAAAA_UAGUUCACC_____ACAUCCCGCAUAGUCUCAGUCUCAGUCCC_AGUCCCCG_C___UCGUCUCCAUUCUCG .......(((((.((..((((((((((...................................)))))..))))))).)))))................................ (-11.73 = -12.17 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:02 2011