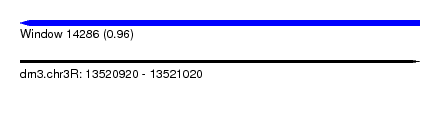
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,520,920 – 13,521,020 |
| Length | 100 |
| Max. P | 0.962507 |

| Location | 13,520,920 – 13,521,020 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.59198 |
| G+C content | 0.59540 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
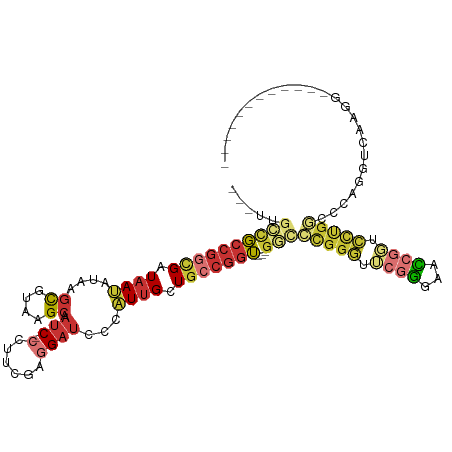
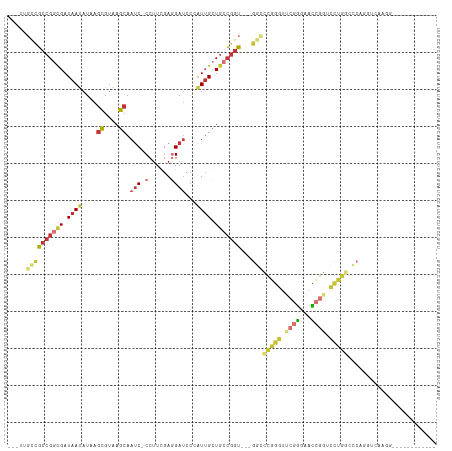
>dm3.chr3R 13520920 100 - 27905053 ---UUGCCGCCGGCGAUAAUAUAAGCGUAAGCAAUC-CCUUCGAGGAUCCCAUUGCUGCCGGUGGCGGCCCGGGUUCGGGAACCGGUCCUGGCCCAGGUCAAGG------------ ---..((((((((((.((((....((....)).(((-(......))))...)))).))))))))))((((.((((.(((((.....))))))))).))))....------------ ( -49.10, z-score = -2.80, R) >droSim1.chr3R_random 725969 101 - 1307089 ---UUGCCGCCGGCGAUAAUAUAAGCGUAAGCAACCUCCUUGGAGGAUCCCAUUGCUGCCGGUGGCGGCCCGGGUUCGGGAACCGGUCCUGGCCCAGGUCAAGG------------ ---..((((((((((.((((....((....))..((((....)))).....)))).))))))))))((((.((((.(((((.....))))))))).))))....------------ ( -53.50, z-score = -3.59, R) >droSec1.super_12 1481612 100 + 2123299 ---UUGCCGCCGGCGAUAAUAUAAGCGUAAGCAAUC-CCUUCGAGGAUCCCAUUGCUGCCGGUGGCGGCCCGGGUUCGGGAACCGGUCCUGGCCCAGGUCAAGG------------ ---..((((((((((.((((....((....)).(((-(......))))...)))).))))))))))((((.((((.(((((.....))))))))).))))....------------ ( -49.10, z-score = -2.80, R) >droYak2.chr3R 2093726 97 - 28832112 ---UUGCCGCCGGCGAUAAUAUAAGUGUAAGCAAUC-CCUUCGAGGACCCCAUUGCUGCCGGU---GGCCCGGGUUCGGGAACCGGUCCUGGCCCAAGUCAAGG------------ ---..(((((((((.....(((....)))(((((((-(......))).....)))))))))))---)))..((((.(((((.....))))))))).........------------ ( -37.10, z-score = -0.82, R) >droEre2.scaffold_4770 9636374 97 + 17746568 ---UUGCCGCCGGCGAUAAUAUAAGCGUAAGCAAUC-CCUUCGAGGAUCCCAUUGCUGCCGGU---GGCCCGGGUUCGGGAACCGGUCCUGGCCCAGGUCAAGG------------ ---..((((((((((.((((....((....)).(((-(......))))...)))).)))))))---)))((((((.(((((.....))))))))).))......------------ ( -42.10, z-score = -1.64, R) >droWil1.scaffold_181108 1523507 87 - 4707319 UUGUCGGUGCCGGUGAUAAUAUAAGCGUUAGCAAUC-CCUUUGAGGAUCCAAUUGCUGGCGGU---GCUGUGGCCCCUGGUGCUGUUGUUG------------------------- ....(((..((((.............((((((((((-(......)))).....)))))))((.---((....))))))))..)))......------------------------- ( -26.70, z-score = -0.61, R) >droVir3.scaffold_12855 10042496 85 - 10161210 ------UUGCCGGUGAUAACAUAAAUGUGAGCAAUC-CCUUUGAAGAUCCUAUUGCUGGCGGU---GUGGCCGCUGGUGGUGUUGCUGUUGCUGC--------------------- ------....(((..((..(((....)))(((((..-((...(.....).....((..((((.---....))))..))))..)))))))..))).--------------------- ( -24.90, z-score = -0.52, R) >anoGam1.chr2R 52274376 109 + 62725911 ------CCUCUGGCGAUAACAUUAGCGUGAGCAAUC-CCUUUGAUGAUCCCGUGGGUCCACAGCGUCCCCCGUAUCAGCAGGGCGCUCCCUAUCCACCGGUUUCACGUCCCCAAGG ------(((.(((.(((.......((....))....-.......(((..((((((((....(((((((..(......)..)))))))....))))).)))..))).))).)))))) ( -36.70, z-score = -2.40, R) >consensus ___UUGCCGCCGGCGAUAAUAUAAGCGUAAGCAAUC_CCUUCGAGGAUCCCAUUGCUGCCGGU___GGCCCGGGUUCGGGAACCGGUCCUGGCCCAGGUCAAGG____________ .....((((.(((((((.......((....)).....((.....)).....))))))).))))...((.(((((.((((...)))).))))).))..................... (-18.10 = -19.10 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:01 2011