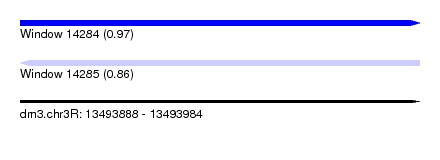
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,493,888 – 13,493,984 |
| Length | 96 |
| Max. P | 0.968454 |

| Location | 13,493,888 – 13,493,984 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Shannon entropy | 0.37643 |
| G+C content | 0.53819 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -32.31 |
| Energy contribution | -32.32 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13493888 96 + 27905053 ----------AUAAGAGUGACAUUCAUGACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAG----- ----------......(((.....)))...((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -33.60, z-score = -1.75, R) >droSim1.chr3R 19531861 96 - 27517382 ----------AAAAGAGGCGUAUUCACGAAGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAG----- ----------........(((....)))..((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -33.60, z-score = -1.77, R) >droSec1.super_12 1454519 96 - 2123299 ----------AAAAGAGGCGUAUUCACGAAGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAG----- ----------........(((....)))..((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -33.60, z-score = -1.77, R) >droYak2.chr3R 2065581 96 + 28832112 ----------AAAAGAGGCGUAUUCAUGACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAG----- ----------....(((.....))).....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -33.30, z-score = -1.48, R) >droEre2.scaffold_4770 9608872 96 - 17746568 ----------AAAAGAGGCGUAUUCACGACGGGGAUGUAGCUCAGAUGGUAGAGCCCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAUU----- ----------......(.(((....))).)((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -32.80, z-score = -1.10, R) >droAna3.scaffold_13340 5785179 96 + 23697760 ----------AAAAAUGGCCAAAUCACGACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAAAU----- ----------....................((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....----- ( -32.90, z-score = -1.91, R) >dp4.chr2 7099356 96 - 30794189 ----------AUAUUGGGCUUAUCCACAACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAA----- ----------...((((((((((((.......))))).)))))))...((.((((....)))).))...((.(((((.(((((.......))))).)))))))...----- ( -35.00, z-score = -1.65, R) >droPer1.super_3 903042 99 - 7375914 ----------AUAUUGGGCUUAUCCACAAGGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAAGGA-- ----------...((((((((((((.......))))).)))))))...((.((((....)))).))...((.(((((.(((((.......))))).)))))))......-- ( -35.00, z-score = -1.14, R) >droWil1.scaffold_181089 8591638 101 + 12369635 ----------AAAUCACAGCAAAUCAUCACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAGGAAGAG ----------.............((.((..((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...)).)). ( -34.20, z-score = -1.71, R) >droVir3.scaffold_13047 2121559 96 - 19223366 ----------AAAUGCUGGGCAUGUACAACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAGGA----- ----------..(((((((((...((((.......))))((((........))))....))))))))).((.(((((.(((((.......))))).)))))))...----- ( -35.90, z-score = -1.32, R) >droMoj3.scaffold_6540 14536968 101 + 34148556 ----------AAAUGCCCAGGAGAUACAACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAGGAUAUUU ----------(((((.((.(......)...((((((((..((((.(((.(.((((....))))).))).))))......((((.......)))))))))))).)).))))) ( -34.70, z-score = -1.65, R) >droGri2.scaffold_14906 2009974 101 - 14172833 ---------AAAAUGC-AGUCGGAUACAACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAGGAUAAAC ---------.......-.((((....)...((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))..))).... ( -33.30, z-score = -1.36, R) >anoGam1.chrU 24927943 110 + 59568033 GGGUGUUACAGAUUGUGCCCCCUAUCACAUGGGGAUGUAGCUCAG-UGGUAGAGCGCUCGCUUCGCAUGUGAGAAGUCCCGGGUUCAAUCCCCGGCAUCUCCAAAAUUCGC (((..(........)..))).........((((((((...((((.-..(..((((....))))..)...)))).....(((((.......)))))))))))))........ ( -36.60, z-score = -0.85, R) >consensus __________AAAUGAGGCGUAUUCACGACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAAA_____ ..............................((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))......... (-32.31 = -32.32 + 0.01)

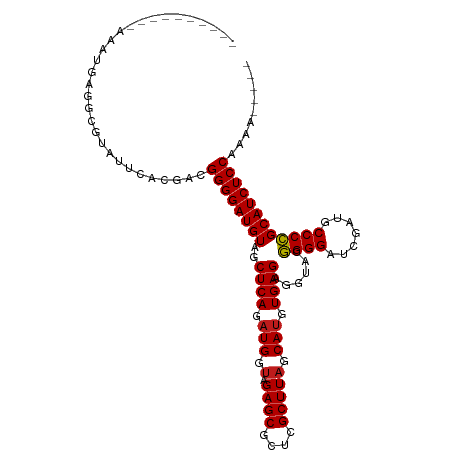
| Location | 13,493,888 – 13,493,984 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Shannon entropy | 0.37643 |
| G+C content | 0.53819 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -22.89 |
| Energy contribution | -22.91 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13493888 96 - 27905053 -----CUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUCAUGAAUGUCACUCUUAU---------- -----....((((.(((((((.......))))))).)))).(((((((.....)))((((........))))................)))).........---------- ( -24.90, z-score = -0.67, R) >droSim1.chr3R 19531861 96 + 27517382 -----CUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCUUCGUGAAUACGCCUCUUUU---------- -----.....(((.(((((((.......))))))).)))((((..(((.....)))((((........))))............)))).............---------- ( -27.50, z-score = -1.77, R) >droSec1.super_12 1454519 96 + 2123299 -----CUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCUUCGUGAAUACGCCUCUUUU---------- -----.....(((.(((((((.......))))))).)))((((..(((.....)))((((........))))............)))).............---------- ( -27.50, z-score = -1.77, R) >droYak2.chr3R 2065581 96 - 28832112 -----CUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUCAUGAAUACGCCUCUUUU---------- -----....((((.(((((((.......))))))).))))..((((((.....)))((((........))))............)))..............---------- ( -24.40, z-score = -0.67, R) >droEre2.scaffold_4770 9608872 96 + 17746568 -----AAUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGGGCUCUACCAUCUGAGCUACAUCCCCGUCGUGAAUACGCCUCUUUU---------- -----....((((.(((((((.......))))))).))))............((((((((........))))...........(((....)))))))....---------- ( -27.10, z-score = -0.60, R) >droAna3.scaffold_13340 5785179 96 - 23697760 -----AUUUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUCGUGAUUUGGCCAUUUUU---------- -----.....(((.(((((((.......))))))).)))((((..(((.....)))((((........))))............)))).............---------- ( -28.30, z-score = -1.28, R) >dp4.chr2 7099356 96 + 30794189 -----UUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUUGUGGAUAAGCCCAAUAU---------- -----..((((((.(((((((.......))))))).)))(((((.(((.....)))((((........))))...........))))).......)))...---------- ( -27.30, z-score = -0.80, R) >droPer1.super_3 903042 99 + 7375914 --UCCUUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCCUUGUGGAUAAGCCCAAUAU---------- --.....((((((.(((((((.......))))))).)))(((((.(((.....)))((((........))))...........))))).......)))...---------- ( -27.30, z-score = -0.97, R) >droWil1.scaffold_181089 8591638 101 - 12369635 CUCUUCCUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGAUGAUUUGCUGUGAUUU---------- ..........(((.(((((((.......))))))).)))(((((.((.........((((........))))..((((.....))))....)))))))...---------- ( -30.50, z-score = -1.56, R) >droVir3.scaffold_13047 2121559 96 + 19223366 -----UCCUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUUGUACAUGCCCAGCAUUU---------- -----....((((.(((((((.......))))))).))))...(((((..(((...((((........))))((((.......))))..)))..)))))..---------- ( -29.90, z-score = -1.75, R) >droMoj3.scaffold_6540 14536968 101 - 34148556 AAAUAUCCUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUUGUAUCUCCUGGGCAUUU---------- ((((..((.((((((((((((.......))))(((...((((.(((.(((((....))).)).))).)))).))).........)))))))).))..))))---------- ( -34.70, z-score = -2.83, R) >droGri2.scaffold_14906 2009974 101 + 14172833 GUUUAUCCUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUUGUAUCCGACU-GCAUUUU--------- (((((.(.(((((((((((((.......)))))))..)))).)).).)))))..((((((........)))).........((((....)))).-)).....--------- ( -29.90, z-score = -1.87, R) >anoGam1.chrU 24927943 110 - 59568033 GCGAAUUUUGGAGAUGCCGGGGAUUGAACCCGGGACUUCUCACAUGCGAAGCGAGCGCUCUACCA-CUGAGCUACAUCCCCAUGUGAUAGGGGGCACAAUCUGUAACACCC ((......(((((((.(((((.......))))).)..)))).))......))..((((((.....-..)))).....((((........))))))................ ( -33.10, z-score = -0.60, R) >consensus _____UUUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUCGUGAAUACGCCUCAUUU__________ .........((((.(((((((.......))))))).)))).....(((.....)))((((........))))....................................... (-22.89 = -22.91 + 0.02)
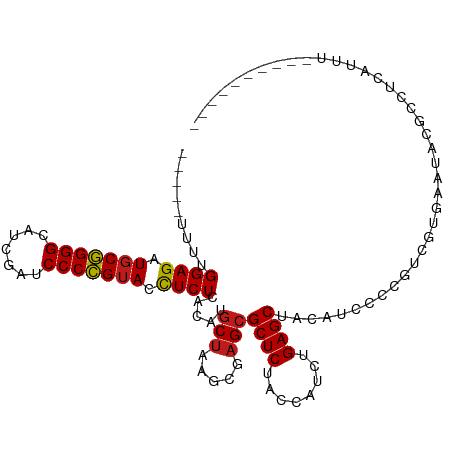
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:23:00 2011